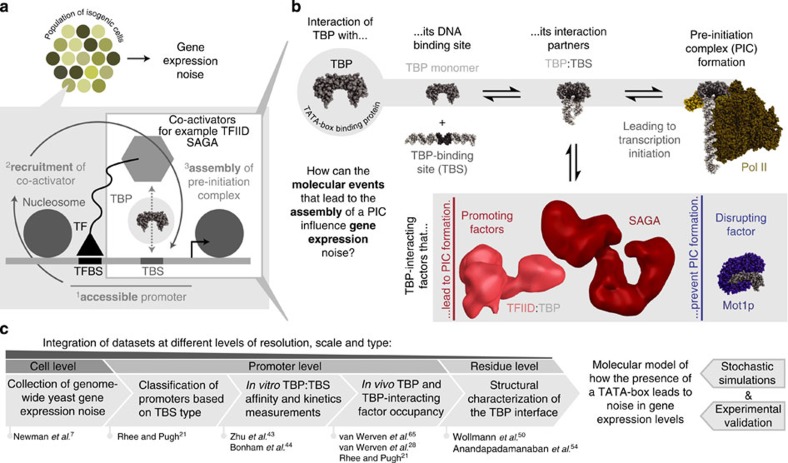
Figure 1. Framework for investigating how gene expression noise arises from the mechanistic details of the interaction of TBP with its partner proteins and their influence on PIC formation.
(a) In a population of genetically identical cells (isogenic population), individual cells can show differences in their gene expression levels (shades of yellow). In order for genes to be expressed, their promoter needs to be accessible (nucleosome re-organization) and co-activating complexes need to be recruited (via transcription factors; TFs). (b) The TATA-box binding protein (TBP) is required for every transcriptional event in eukaryotic cells. TBP can exist in different functional assemblies. They are in dynamic equilibrium between a dimeric state and a monomeric state that in turn can form different TBP assemblies with (i) other general transcription factors (TFIIB and TFIIA in yellow), (ii) co-activators (TFIID in pink and SAGA in red) that promote pre-initiation complex (PIC) formation or (iii) disrupting factors (Mot1p in purple) that bind and evict the DNA bound TBP and prevent PIC formation. (c) To obtain mechanistic and molecular insights into the origins of noise, we integrated data from different levels of resolution, scales and types describing various aspects of transcription initiation in the yeast Saccharomyces cerevisiae, which were tested using stochastic simulations and were experimentally validated.