Fig. 1.
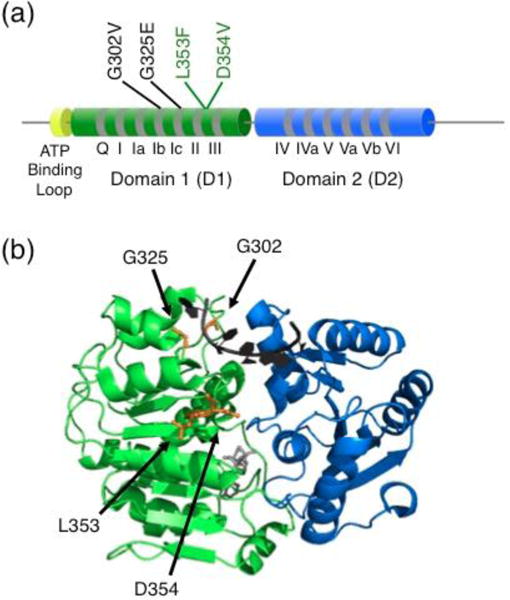
Medulloblastoma-linked DDX3X mutations studied in vitro and in S. pombe. (a) Schematic with domain architecture of DDX3X. Mutations that resulted in compromised RNA-dependent ATPase activity and failed to complement a lack of functional Ded1 in S. pombe are labeled in black. Mutations that did not reveal detectable defects are labeled in green. The study included five additional mutations, not shown, that were probed in vivo but could not be purified in soluble form to study in vitro. The newly-revealed ATP-binding loop is shown in yellow. The cylinders and line connecters are not to scale. (b) Approximate locations of mutations in the closed core complex. The structure depicts D1 and D2 of Vasa [28], and the amino acids that are homologous to those mutated by Epling et al are colored orange and labeled with DDX3X numbering. Bound ssRNA is shown in black and bound AMP-PNP is shown in gray.
