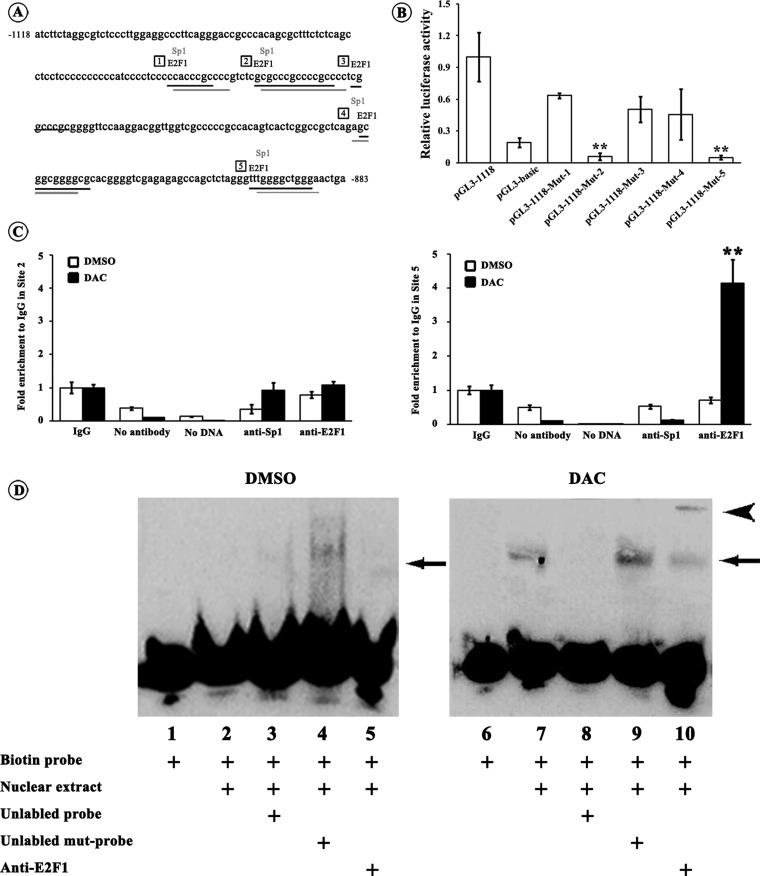
Figure 3. Identification of transcription factor binding sites in miR-34c promoter.
(A) Sequence of the miR-34c promoter flanking from −1118 to −1381 bp and potential E2F1- and Sp1-binding sites within it. E2F1 and Sp1 shares common binding sites in the miR-34c promoter; and we set Site 1–Site 5 for site-directed mutagenesis assays. Numbers in frames indicates the five binding sites. (B) Each pGL3-1118 mutant (Mut-1–Mut-5) luciferase reporter construct or pGL3-1118 construct was transfected into 293T cells. Twenty-four hours after the transfection, the dual luciferase assays were performed. When compared with pGL3-1118 construct, the relative luciferase activities of pGL3-1118-Mut-2 and pGL3-1118-Mut-5 constructs were significantly lowered to a level of pGL3-basic plasmid. All luciferase assays were performed in six times (**P<0.01). (C) HT-29 cells were treated with DAC or DMSO followed by ChIP assays. Chromatin was immunoprecipitated with the anti-E2F1 or anti-Sp1 antibody. The real-time PCR specific to Site 2 and Site 5 were performed. For the control, no antibody or no DNA was used. The values were shown relative to IgG group which was set to 1. Only the binding activity of E2F1 to Site 5 was evidently enhanced upon the DAC treatment. All PCR reactions were performed in triplicates (**P<0.01). (D) Nuclear extract from HT-29 cells treated with DAC or DMSO were incubated with biotin-labelled probe corresponding to putative E2F1 binding Site 5 in the miR-34c promoter (lanes 2–5 and 7–10). For competition system, 200-fold excess of unlabelled probe or unlabelled mutational probe was additionally added (lanes 3, 4, 8 and 9). For supershift assay, anti-E2F1 antibody was included (lanes 5 and 10). The negative control was used in the absence of nuclear extracts (lanes 1 and 6). It clearly showed the specific binding of E2F1 to the Site 5 (−897 to −889 bp) in the miR-34c promoter after DAC induced demethylation. The arrow indicates the specific DNA–protein complex and the arrowhead indicates the supershift.