Short insertions/deletions in homeolog genes affect microRNA-mediated posttranscriptional regulation and are important for stresses like viral infections that alter the expression of microRNAs.
Abstract
The plant ARGONAUTE1 protein (AGO1) is a central functional component of the posttranscriptional regulation of gene expression and the RNA silencing-based antiviral defense. By genomic and molecular approaches, we here reveal the presence of two homeologs of the AGO1-like gene in Nicotiana benthamiana, NbAGO1-1H and NbAGO1-1L. Both homeologs retain the capacity to transcribe messenger RNAs (mRNAs), which mainly differ in one 18-nucleotide insertion/deletion (indel). The indel does not modify the frame of the open reading frame, and it is located eight nucleotides upstream of the target site of a microRNA, miR168, which is an important modulator of AGO1 expression. We demonstrate that there is a differential accumulation of the two NbAGO1-1 homeolog mRNAs at conditions where miR168 is up-regulated, such as during a tombusvirus infection. The data reported suggest that the indel affects the miR168-guided regulation of NbAGO1 mRNA. The two AGO1 homeologs show full functionality in reconstituted, catalytically active RNA-induced silencing complexes following the incorporation of small interfering RNAs. Virus-induced gene silencing experiments suggest a specific involvement of the NbAGO1 homeologs in symptom development. The results provide an example of the diversity of microRNA target regions in NbAGO1 homeolog genes, which has important implications for improving resilience measures of the plant during viral infections.
The ARGONAUTE1 gene (AGO1) was first discovered in Arabidopsis (Arabidopsis thaliana) and further described trough plant mutants that showed pleiotropic developmental anomalies. Examples of phenotypic traits influenced by AGO1 are cotyledon expansion, leaf polarity, branching of the inflorescence stem, rooting, differentiation of flowers, and fertility (Kidner and Martienssen, 2004, 2005; Vaucheret et al., 2004; Sorin et al., 2005). The pleiotropic character of AGO1 is determined by the role of the AGO1 protein as a core component of the microRNA (miRNA)/RNA-induced silencing complex (RISC) in posttranscriptional gene silencing (PTGS; Baumberger and Baulcombe, 2005). Most miRNAs are incorporated into AGO1 and guide the RISC to its mRNA target through sequence complementarity; as a result, the mRNA translation is inhibited by endonucleolytic cleavage or other yet incompletely characterized mechanisms (Brodersen et al., 2008; Lanet et al., 2009; Iwakawa and Tomari, 2013). Beside its role in PTGS and in concert with other proteins of the Argonaute clade, AGO1 plays a key role in the RNA silencing-based antiviral defense (Takeda et al., 2008; Harvey et al., 2011; Scholthof et al., 2011a; Wang et al., 2011); similar to miRNAs, virus-derived small interfering RNAs (vsiRNAs) are incorporated into AGO1-containing RISC, leading to the inactivation of viral RNAs by cleavage. In turn, viruses may encode viral suppressors that impair AGO1 functionality (for review, see Burgyán and Havelda, 2011).
AGO1 homeostasis is in part coordinated through a feedback mechanism. Thus, microRNA168 (miR168)-guided cleavage of AGO1 mRNA was demonstrated to ensure an optimal balance of miRNA steady-state levels (Vaucheret et al., 2004, 2006; Vazquez et al., 2004; Vaucheret, 2009). Conversely, overexpression of miR168 (e.g. in plants expressing miR168 under the control of the 35S promoter) is accompanied by a decrease in AGO1 mRNA accumulation, and it is associated with the cleavage of AGO1 mRNA. Moreover, miR168 also overaccumulates in plants that express a miR168-resistent AGO1 mRNA (i.e. 4m-AGO1), owing to stabilization by AGO1 (Vaucheret et al., 2004, 2006).
Nicotiana benthamiana is one of the most widely used models to study RNA virus-plant interactions. Upon infection, N. benthamiana activates and orchestrates an antiviral RNA interference response, which mostly parallels the RNA interference pathways that were described for the model plant Arabidopsis (Qiu et al., 2002; Omarov et al., 2006; Pantaleo et al., 2007; Goodin et al., 2008; Csorba et al., 2009; Várallyay et al., 2010; Scholthof et al., 2011a, 2011b). The recent release of the draft genome sequence of N. benthamiana provides the opportunity to shed new light on additional layers of plant-virus interaction, because N. benthamiana is susceptible to a considerably broader spectrum of viruses than Arabidopsis.
Tombusviruses like Cymbidium ringspot virus (CymRSV) and Tomato bushy stunt virus (TBSV) are well-accepted prototypes to study RNA silencing-based plant-virus interactions. Tombusviruses have a broad host range spanning approximately 20 plant families and approximately 120 species (Yamamura and Scholthof, 2005). The tombusvirus genomic and subgenomic RNAs are substrates of DICERS yielding high levels of viral small interfering RNAs (siRNAs; Szittya et al., 2002, 2010), and these viruses encode a 19-kD protein (P19) that is a potent suppressor of RNA silencing (Voinnet et al., 1999; Vargason et al., 2003). While Arabidopsis is not permissive to a tombusvirus infection, CymRSV and TBSV replicate in heterologous systems such as Saccharomyces cerevisiae (Panavas and Nagy, 2003; Pantaleo et al., 2003; Navarro et al., 2006). TBSV also was recently applied in a plant cytoplasm-based system to reproduce viral replication as well as antiviral RNA silencing in vitro (Gursinsky et al., 2009; Schuck et al., 2013).
Nicotiana spp. frequently contain so-called homeologs; these are duplicated genes in the same plant genome derived from different ancestors in polyploidy (Bombarely et al., 2012a). In this report, we show that N. benthamiana possesses two NbAGO1-like homeolog genes, NbAGO1-1H and NbAGO1-1L. They differ mainly by one 18-nucleotide-long insertion/deletion (indel), which does not modify the translational frame. Importantly, the indel is located in the immediate vicinity of the miR168 target site, and our data suggest that its presence considerably affects the miR168-guided posttranscriptional regulation of NbAGO1 mRNA. The indel effect is highlighted under conditions of an increased miR168 accumulation (e.g. during a viral infection). The two NbAGO1 homeologs show full functionality in reconstituted, catalytically active RISC following the incorporation of siRNAs. Moreover, virus-induced gene silencing (VIGS) experiments suggest a specific, redundant involvement of the NbAGO1 homeologs in susceptibility to viral infection but a divergent involvement in symptom development. The expression of two types of NbAGO1-1 mRNAs is proposed to represent an evolutionary adaptation to improve resilience measures of the plant during viral infections or other stresses accompanied by miR168 up-regulation.
RESULTS
A New AGO1 Locus in the N. benthamiana Genome
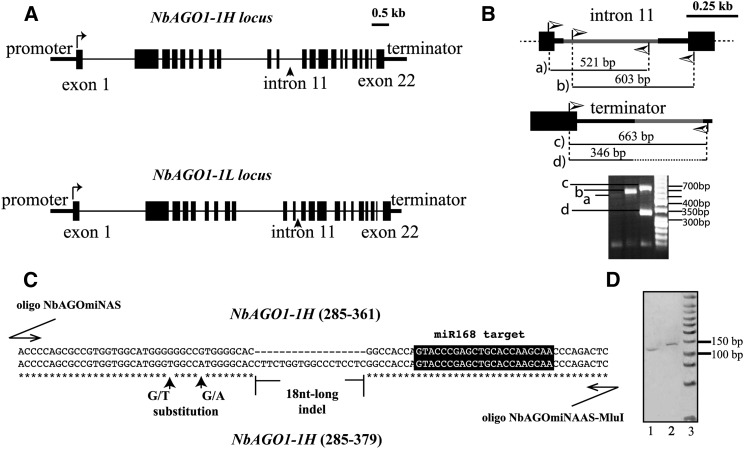
Previously, two AGO1-like genes were identified in N. benthamiana: NbAGO1-1 and NbAGO1-2 (National Center for Biotechnology Information accession nos. DQ321488 and DQ321489, respectively). Both NbAGO1-like genes were shown to be required for a full systemic silencing of transgenes, which is in close analogy to observations made with hypomorphic mutants of Arabidopsis AGO1 alleles (Jones et al., 2006). We retrieved the NbAGO1 genes from the N. benthamiana draft genome available for public research (http://solgenomics.net/organism/Nicotiana_benthamiana/genome) and, unexpectedly, identified an additional NbAGO1 locus. By comparing the sequence of the second exon, we determined a high similarity with the 5′-terminal sequence of NbAGO1-1 (DQ321488) but not with NbAGO1-2 (DQ321489). A pairwise alignment of the known and the newly determined NbAGO1-1 loci revealed a different length; accordingly, we renamed the two loci as NbAGO1-1L (for low) and NbAGO1-1H (for high). The NbAGO1-1L locus (corresponding to DQ321488) is 7,008 bp long, while the NbAGO1-1H locus is 7,195 bp long (introns and exons included but promoter and terminator sequences excluded; Supplemental Fig. S1). Both loci contain 22 exons and 22 introns, and the different length is mainly explained by indels at the intron level (e.g. within intron 11; Fig. 1A; Supplemental Fig. S1).
Figure 1.
Organization of NbAGO1-1H and NbAGO1-1L loci. A, Schematic representation of the structures of NbAGO1-1H (top) and NbAGO1-1L (bottom) loci. Black lines represent introns, vertical bars represent exons, and horizontal bars at the two extremities are untranslated regions. B, Graphic representing the PCR-based approach used to reveal the two alternative NbAGO1-1 loci in the intron 11 and terminator regions (top and middle, respectively). The high-resolution gel electrophoresis of the PCR products is reported at the bottom. Lowercase letters (i.e. a–d) indicate single amplification products, which are represented as thin black lines at the top and middle. C, Alignment of the section of exon 2 containing the 18-nucleotide-long indel, which was used to reveal the expression of the two NbAGO1-1 transcripts. The applied oligonucleotides are described in “Materials and Methods.” Stars indicate the consensus sequence; black vertical arrows mark two detected nucleotide substitutions. The black box highlights the miR168 target site. D, High-resolution acrylamide gel electrophoresis of the two RT-PCR fragments from the two RNA transcripts described in C. Lanes 1 and 2 show the 129- and 147-bp-long amplicons that were obtained from the NbAGO1-1L and NbAGO1-1H transcripts, respectively. A low Mr New England Biolabs marker is in lane 3.
To validate the gene sequences that were retrieved from the N. benthamiana genomic data, we searched for intronic/exonic sequences to design and apply a PCR-based strategy to specifically discriminate between the two NbAGO1-1 loci. Thus, specific oligonucleotides were designed that annealed exclusively within the NbAGO1-1H intron 11 (gray bar in Fig. 1B, top; Supplemental Fig. S2A). We applied a similar approach to the terminator region, where an approximately 300-bp-long indel discriminates the two NbAGO1-1 loci (gray bar in Fig. 1B, middle; Supplemental Fig. S2B). The obtained PCR products had the expected sizes (PCR amplicons in Fig. 1B, bottom); they were subsequently cloned and sequenced. The obtained sequence data confirmed the N. benthamiana draft genome sequence (Bombarely et al., 2012b).
NbAGO1-1L and NbAGO1-1H Are Functional in Generating Transcripts
An 18-nucleotide-long indel located within exon 2 (position 322, referred to as the coding sequence section; Supplemental Fig. S1) was used to detect and discriminate NbAGO1-1L and NbAGO1-1H transcripts in N. benthamiana. First, polyadenylated RNAs were selectively purified from total RNA and randomly reverse transcribed. The complementary DNA (cDNA) was then subjected to PCR using oligonucleotides that were applicable to both transcripts of NbAGO1-1L and NbAGO1-1H encompassing the 18-nucleotide-long indel (see “Materials and Methods” and alignment in Fig. 1C). The PCR was first analyzed by high-resolution gel electrophoresis. It revealed two main amplicon species, which were gel purified and again resolved by gel electrophoresis. In Figure 1D, ethidium bromide-stained amplicons are shown to have the expected (i.e. based on the retrieved sequence) lengths of 129 bp (from the NbAGO1-1L transcript) and 147 bp (from the NbAGO1-1H transcript; lanes 1 and 2, respectively). Sanger sequencing of the two amplicons confirmed the presence of both the indel and of certain G/T and T/A polymorphic nucleotides (single-nucleotide polymorphisms; Fig. 1C). The characteristics of both transcripts were further confirmed through the amplification and sequencing of longer segments (data not shown).
Recently, an N. benthamiana transcriptome analysis was carried out by RNA deep sequencing and assembly (Nakasugi et al., 2013). The available data set was searched for the NbAGO1-1 transcripts and, indeed, both species were found. Based on the 18-nucleotide-long indel (Fig. 1C), Nbv3K765634670 is the sole transcript that represents NbAGO1-1L, whereas several other transcripts represent NbAGO1-1H (i.e. Nbv3K605752598, Nbv3K805664652, Nbv3K705828682, Nbv3K705826800, Nbv3K745621734, and Nbv3K745621399; Supplemental Fig. S3). The study of Nakasugi et al. (2013) did not highlight the presence of the NbAGO1-1L transcript (see “Discussion”).
The miR168-Guided Silencing Machinery Cleaves NbAGO1-1H and NbAGO1-1L at the Target Site
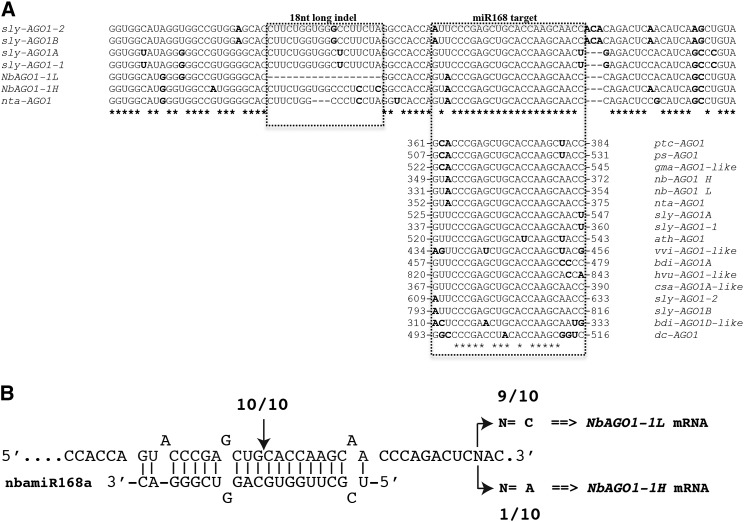
miR168 is a highly conserved miRNA within the plant kingdom (Supplemental Fig. S4; Cuperus et al., 2011). Accordingly, the miR168 target site is also highly conserved within the annotated NbAGO1 sequences (Fig. 2A). Both NbAGO1-1L and NbAGO1-1H possess an identical putative miR168 target site and, notably, the 18-nucleotide-long indel is located only eight nucleotides upstream (Fig. 2A). In order to prove whether miR168 targets and cleaves the NbAGO1-1 mRNAs in N. benthamiana, we performed a 5′-RACE analysis. Ten clones out of 10 sequences confirmed the expected cleavage between nucleotides complementary to positions 10 and 11 of miR168 (Fig. 2B, vertical arrow). These data accordingly recapitulate the miR168-driven cleavage of Arabidopsis AGO1 mRNA (Rhoades et al., 2002; Vaucheret et al., 2004).
Figure 2.
Sequence diversity of the indel-containing region and the miR168 target. A, Sequence diversity of the indel-containing region of all known annotated or predicted AGO1 mRNAs from plants belonging to the Solanaceae family. Highlighted are the 18-nucleotide-long indel (box on the left) and the miR168 target site (box on the right). In the latter case, the alignment analysis was extended to other plant species. Sequences in boldface are not conserved and show a nucleotide occurrence of less than 50%. B, 5′-RACE analysis of the miR168-mediated cleavage site. The vertical arrow indicates the positions corresponding to the 5′ ends of NbAGO1-1L and NbAGO1-1H RNAs that were determined by 5′-RACE and the number of 5′-RACE clones corresponding to the site. The single-nucleotide polymorphism (indicated by N) at the 3′ end of the reported sequence was used as a molecular mark to discriminate the cleaved alternative transcript. Accession numbers of retrieved annotated or predicted AGO1 mRNAs are as follows: ptc, Populus trichocarpa (XM002318302); ps, Pisum sativum (EF108450); gma, Glycine max (XM003548263); nta, N. tabacum (AB542739); sly, S. lycopersicum (JX467704, JX945381, JX945382, and JX467705); ath, Arabidopsis (NM001198240); vvi, Vitis vinifera (XM002271189); bdi, Brachypodium distachyon (XM003580292 and XM_003563186); hvu, Hordeum vulgare (AK373112); csa, Cucumis sativus (XM004137339); and dc, Daucus carota (AB360853).
Taking advantage of the presence of a polymorphic nucleotide at position 380 (Fig. 2B, base N), which is located eight nucleotides downstream of the miR168 target sequence, we could discriminate the miR168-driven cleavage events for NbAGO1-1L and NbAGO1-1H (i.e. C and A, respectively, in Fig. 2A). Thus, the 5′ RACE revealed a proportion of one H species (characterized by the A nucleotide) versus nine L species (characterized by the C nucleotide; Fig. 2B). These data suggest that both NbAGO1-1 transcripts were targeted and cleaved by the miRNA-mediated silencing machinery; however, the cleavage remnants that were identified by the 5′-RACE analysis (see above) more frequently derived from the L homeolog.
The 18-Nucleotide-Long Indel Affects the miR168-Mediated Expression of a GFP Reporter Gene
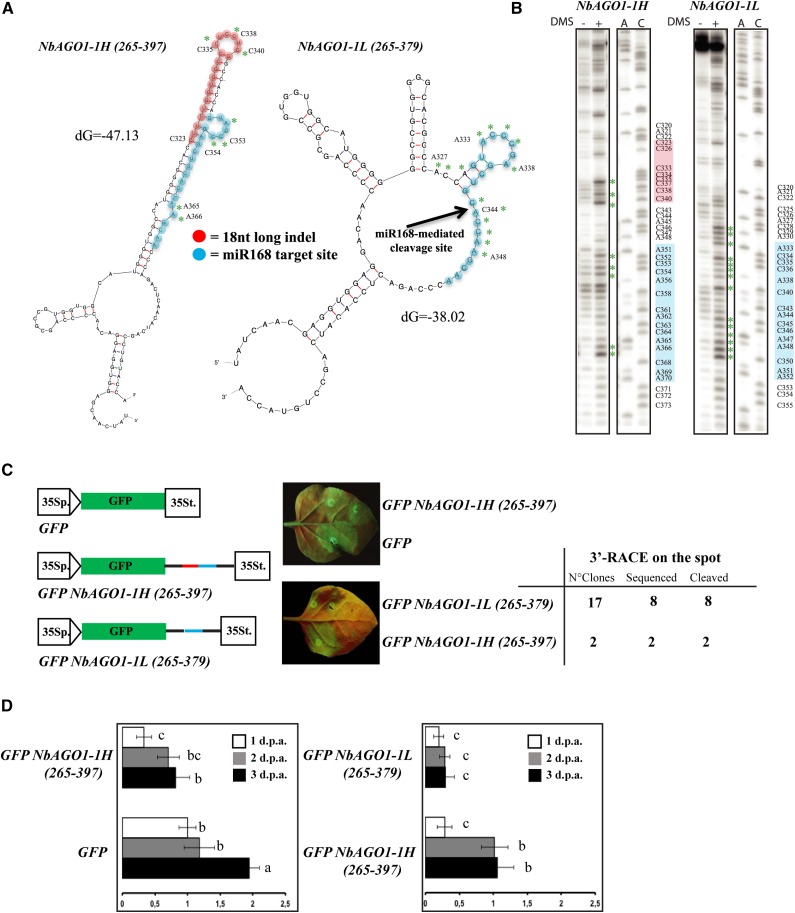
It was previously shown in vivo as well as in vitro that the accessibility of small RNA target sites directly correlates with target recognition and with the efficiency of cleavage, thus indicating that RISC is unable to unfold structured RNA of both endogenous or exogenous origin (i.e. mRNAs or viral RNAs, respectively; Ameres et al., 2007; Long et al., 2007; Schuck et al., 2013). This has been shown most convincingly in animal systems, but it is widely accepted also in plant systems for antiviral RISC that likely involve components similar to those involved in miRNA-mediated RNA silencing (e.g. AGO1; for review, see Ding and Voinnet, 2007). The 18-nucleotide-long indel is GC rich (12 nucleotides out of 18; it contains eight Cs, four Gs, and six Us), and its close proximity to the miR168 target region (Fig. 2A) prompted us to examine the secondary structure of the region in the NbAGO1-1H and NbAGO1-1L context. Therefore, we submitted two stretches of sequence from positions 265 to 378 (for NbAGO1-1L) and from positions 265 to 396 (for NbAGO1-1H) to Mfold (Zuker, 2003; Supplemental Fig. S1, coding sequence section). The output of the program is shown in Figure 3A. It suggested that the NbAGO1-1H (265-396) RNA is stably structured in this region and that the miR168 target sequence is masked by endogenous base pairings (Fig. 3A, left). In contrast, in the NbAGO1-1L (265-378) RNA, the miR168-binding site was suggested to be less structured; accordingly, it seems more accessible to miRNA-directed targeting (Fig. 3A, left versus right).
Figure 3.
miR168-resistent target site in NbAGO1-1. A, Mfold output showing the secondary structures of NbAGO1-1H (left) and NbAGO1-1L (right) in the region containing the miR168 target site. B, Chemical probing (DMS) of RNA secondary structure and primer extension analysis. Green asterisks in A and B indicate modified bases. C, In vivo analysis of GFP NbAGO1-1 sensor sequences. Spots in leaf halves were infiltrated with A. tumefaciens containing the indicated sensor sequences. The miR168 target site sequence is shown in red; the differential 18-nucleotide-long indel is shown in blue. At right is a table showing the 3′-RACE analysis on the spot. D, Relative expression levels of GFP mRNA in the analysis of GFP NbAGO1-1 sensor sequences. Data are presented as means ± se of three replicates; different letters denote significant differences at P ≤ 0.05. GFP mRNA at 1 dpa = 1.
To experimentally verify the predicted Mfold structures, we performed chemical modification experiments. Thus, transcripts NbAGO1-1L (265-378) and NbAGO1-1H (265-396) were generated by T7 polymerase-driven in vitro transcription from appropriate PCR amplicons and treated with dimethyl sulfate (DMS; see “Materials and Methods”). DMS specifically modifies (methylates) unpaired A and C nucleotides. The sites of chemical modification were subsequently determined by primer extension (see “Materials and Methods”); the identification of the modified nucleotides was enabled by a side-by-side electrophoresis of a DNA sequencing reaction. Figure 3B shows the experiment-deduced RNA structure, where the DMS-modified As and Cs are highlighted by green asterisks. In sum, the experimental data were in robust agreement with the obtained Mfold predictions (compare nucleotides marked by green asterisks in Fig. 3, A and B). Most interestingly, the structures of both transcripts differed considerably. That is, the transcript NbAGO1-1L (265-378) turned out to be significantly more accessible to chemical modifications, particularly the region that represents the target site of the miR168 seed sequence (Figs. 2B and 3B; see “Discussion”).
Next, we asked if the 18-nucleotide indel and its predicted impact on the mRNA’s secondary structures affected miR168-directed PTGS. For this purpose, we adopted a strategy that was earlier applied to monitor the miR171 cleavage of target RNA in planta (Parizotto et al., 2004; Lakatos et al., 2006). Binary vectors were generated that enabled Agrobacterium tumefaciens-mediated transient expression of two alternative reporter mRNAs via the 35S promoter in N. benthamiana (Fig. 3C). Each of these mRNAs consisted of the open reading frame encoding the GFP sensor and a flanking 3′ untranslated region, which contained either the H or the L sequence elements (Fig. 3A). The reporter sequences were transiently coexpressed with the viral suppressor P19 in order to prevent any transgene silencing and to isolate the effect of the plant RISC loaded with the endogenous miRNAs (Lakatos et al., 2006). Visual inspection by UV light revealed that the expressed GFP from GFP NbAGO1-1L (256-378) was barely detectable at 3 d post agroinfiltration (dpa). In contrast, expression of the reporter was high in leaves that were infiltrated with GFP NbAGO1-1H (256-396) and with the negative control (no target (Fig. 3C). This trend was confirmed when we quantified the amounts of the GFP sensor mRNAs at 1, 2, and 3 dpa by quantitative reverse transcription (qRT)-PCR (Fig. 3D). In sum, both sets of data revealed that, in the agroinfiltrated leaves at the indicated time points, the GFP NbAGO1-1L (256-378) mRNA showed a lower level of expression than the GFP NbAGO1-1H (256-396) version (Fig. 3, C and D). Importantly, when we performed a 3′-RACE analysis of the GFP reporter RNAs that were extracted from the agroinfiltrated spots, we obtained two colonies for GFP NbAGO1-1L (256-378) and 17 colonies for GFP NbAGO1-1H (256-396). All the sequences revealed miR168-directed cleavage, which was congruent to the data that were obtained earlier with the NbAGO1-1L and NbAGO1-1H transcripts in planta (Figs. 2B and 3C). These observations provided further direct evidence that the strong down-regulation of the reporter expression, which carried the miR168 target region in the NbAGO1-1L context, was caused by an efficient miR168-driven cleavage.
NbAGO1-1L Is Underrepresented in the Course of a CymRSV Infection
Plant virus infections correlate with a significant accumulation of miR168 in infected tissues (Zhang et al., 2006; Csorba et al., 2007; Havelda et al., 2008; Várallyay et al., 2010, 2014; Lang et al., 2011). The reasons leading to this phenomenon are yet uncertain. They may be attributed either to a direct induction of miR168 expression (e.g. by viral components such as viral suppressors; Várallyay and Havelda, 2013) or to an induction of AGO1 mRNA expression, which, in turn, may lead to a stabilization of miR168 via incorporation into the AGO1 protein (Vaucheret et al., 2006).
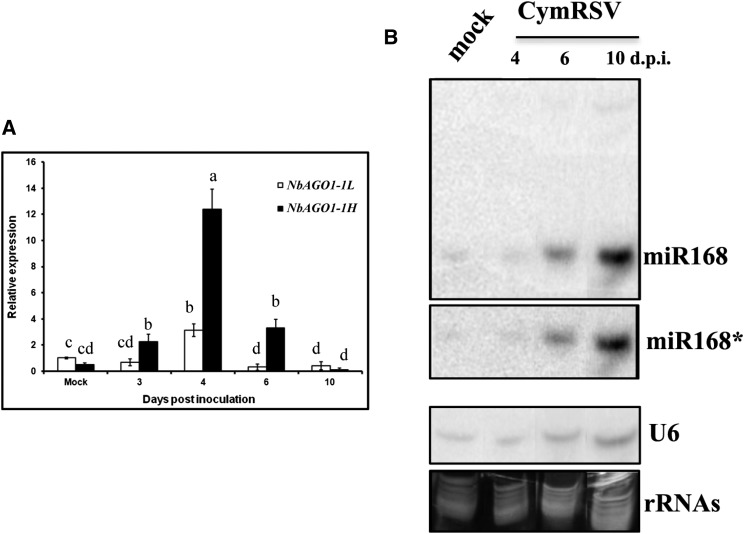
To gain further insights into the miR168-mediated posttranscriptional control of NbAGO1-1 mRNAs, we used the well-characterized CymRSV/N. benthamiana system. Taking advantage of the 18-nucleotide-long indel as the main sequence difference between the two homeolog mRNAs, specific oligonucleotides were designed to enable a fine discrimination between the NbAGO1-1L and NbAGO1-1H transcripts (procedure reported in Supplemental Fig. S5). Measuring the levels of the NbAGO1-1 mRNAs by qRT-PCR, we observed that in the course of a CymRSV infection, the amounts of NbAGO1-1 mRNAs increased dramatically up to 4 d post inoculation (dpi; Fig. 4A), which is in line with earlier observations by Várallyay et al. (2010). However, when discriminating NbAGO1-1L from NbAGO1-1H, the scenario revealed other valuable details (Fig. 4A). Initially, in mock-inoculated plants, the ratio between NbAGO1-1L and NbAGO1-1H mRNAs was about 2:1. Conversely, at 3 dpi, the L:H ratio was inverted to 1:4. As explained, the levels of both H and L mRNAs continued to rise up to 4 dpi, although the ratio remained unaltered. At 6 dpi, the NbAGO1-1L transcripts reached a level close to 0, while the level of the NbAGO1-1H transcripts still was 8 times higher than in the mock-inoculated plants. At 10 dpi, the levels of both transcripts approximated 0, most likely because all tissues of the infected plants were already necrotic. Thus, during a viral infection that induces the accumulation of NbAGO1-1 mRNAs and that is also accompanied by miR168 up-regulation (Vaucheret et al., 2004), the amount of the NbAGO1-1H variant was found to be particularly increased (Fig. 4). These data are in close agreement with the earlier observed frequencies of transcripts and miR168-mediated cleavage products (see above).
Figure 4.
Relative expression of NbAGO1-1L and NbAGO1-1H during CymRSV infection. A, qRT-PCR of the two alternative transcripts; amplification was normalized to CPH transcripts. Data are presented as means ± se of three replicates; different letters denote significant differences at P ≤ 0.05. B, Northern-blot analysis of miR168 and miR168* during CymRSV infection. rRNA, Ribosomal RNA.
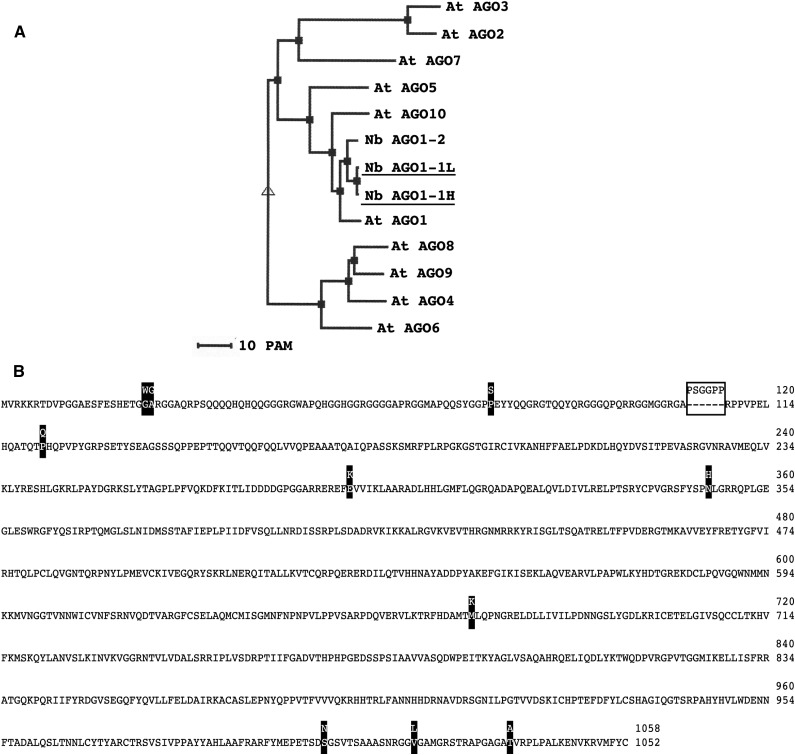
Both NbAGO1 Homeologs Are Catalytically Active
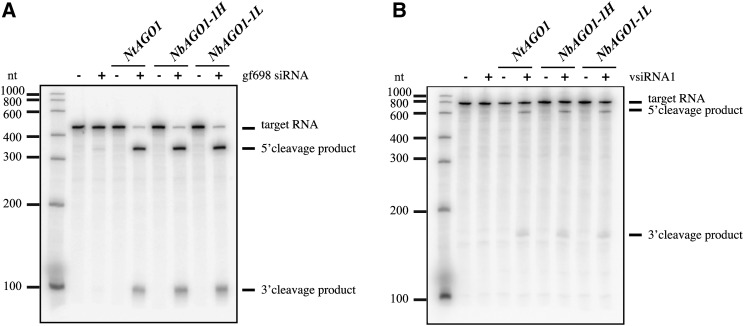
Based on the transcript sequences, putative AGO1 H and L proteins were deduced and aligned using ClustalW (Thompson et al., 1994). A subsequent phylogenetic inspection of Arabidopsis AGO proteins using MultiAlin (http://multalin.toulouse.inra.fr/multalin/) confirmed that the two putative proteins are strongly related and that each belongs to the Arabidopsis AGO1 clade (Vaucheret, 2008; Fig. 5A): in the scheme shown in Figure 5B, amino acid substitutions are highlighted as black boxes while the indel is indicated by the white box. Considering that differences in the N-terminal sequence of human AGO1 were earlier indicated to tune the catalytic activity of the protein (Hauptmann et al., 2013), this prompted us to determine whether the two AGO homeologs are functional (i.e. whether they display a comparable AGO/RISC-mediated RNA cleavage [slicer] activity). However, the evaluation of the biological relevance of the two AGO1 homeologs in planta is difficult, mostly because a differential functional characterization of the H and L AGO1 proteins is impeded by their close similarity (Fig. 5B). For these reasons, we decided to perform this set of experiments in an in vitro system. This system, which is based on cytoplasmic extracts of Nicotiana tabacum BY-2 cells (BY-2 cell lysate [BYL]), is capable of reconstituting active RISC with in vitro-translated Arabidopsis, N. benthamiana, or N. tabacum AGO1 proteins and effectively recapitulates small RNA-driven mRNA regulation (Iki et al., 2010) as well as vsiRNA-driven antiviral silencing (Schuck et al., 2013). In the experiments performed, NbAGO1-1H and NbAGO1-1L cDNAs were in vitro transcribed and the corresponding proteins produced by in vitro translation in the BYL. Importantly, the translation reaction was carried out in the absence and presence of two types of siRNAs, siRNA gf698 and vsiRNA1. These siRNAs were earlier demonstrated to program reconstituted AGO1/RISC such that a GFP-encoding target mRNA and a viral RNA (a TBSV defective interfering RNA) are efficiently targeted and cleaved, respectively (Iki et al., 2010; Schuck et al., 2013). As shown in Figure 6, both types of RISC reconstituted with either the NbAGO1-1H or NbAGO1-1L protein efficiently cleaved their respective target RNA dependent on the programming siRNA. Thus, in comparison with the control reaction, 5′ and 3′ cleavage products were clearly detectable (Fig. 6). Accordingly, we concluded that both NbAGO1-1 homeologs effectively incorporated siRNAs and formed a RISC that was able to slice mRNA and viral RNA targets, respectively.
Figure 5.
NbAGO1-1 proteins. A, Phylogenetic classification of N. benthamiana AGO1 and the Arabidopsis AGO proteins depicted in three clades. PAM indicates the point-accepted mutation. B, NbAGO1-1 predicted protein sequence showing amino acid differences between the H and L forms.
Figure 6.
In vitro slicer activity of NbAGO1-1H and NbAGO1-1L. A, NbAGO1-1 mRNAs were translated in N. tabacum BYL in the absence or presence of an exogenous siRNA (gf698) targeting the mRNA of GFP (Iki et al., 2010). Subsequently, a 32P-labeled GFP mRNA fragment was added as a target, and RISC cleavage products were analyzed by denaturing PAGE and autoradiography. As negative and positive controls, the reactions were carried out in the absence of additionally expressed (in vitro-translated) NbAGO1 and with NtAGO1, respectively. B, The assay was performed as described above except that a TBSV defective interfering RNA was targeted by a vsiRNA, vsiRNA1 (Schuck et al., 2013).
VIGS of NbAGO1-1H, Viral Accumulation, and Silencing of Endogenous Genes
The genetic dissection of homeolog genes that differ only in a few stretches of sequences in N. benthamiana is not an easy task, and it is further complicated by the fact that AGO1 is essentially involved in basic functions of plant development (Vaucheret, 2008). However, it is possible to silence Argonaute genes in N. benthamiana by VIGS via a tobacco rattle virus vector (Jones et al., 2006). Considering a similar approach, we took advantage of the study of Pignatta et al. (2007), who developed a TBSV-based vector. Thus, as a first approach, we inoculated N. benthamiana plants with in vitro transcripts of an empty TBSV (control) and a TBSV:NbAGO1-1 vector (Fig. 7A; for details, see “Materials and Methods”). Both sets of infected plants started to show typical symptoms of a tombusvirus infection, including necrosis in inoculated and systemic leaves at 3 to 4 dpi. However, in contrast to the control plants, the plants that were infected by TBSV:NbAGO1-1 rapidly showed necrotic symptoms leading to a quick decline until death at 10 dpi (Fig. 7A). The increased sensitivity of the plant to viral infection as a consequence of the silencing of NbAGO1-1 is in line with a previous report (Jones et al., 2006) and emphasizes the central role of AGO1 in RNA silencing-based antiviral defense (for review, see Burgyán and Havelda, 2011).
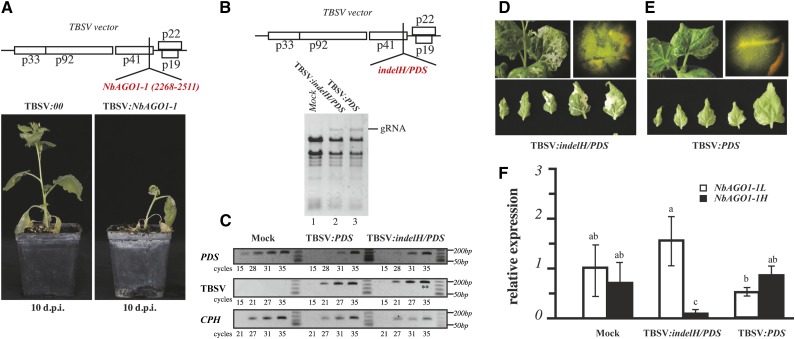
Figure 7.
VIGS of NbAGO1-1H. A, Schematic representation of the TBSV viral vector containing a fragment inducing the silencing of NbAGO1-1 transcripts and the effect of viral vectors on N. benthamiana plants at 10 dpi. B, Schematic representation of the TBSV viral vector containing the fragment that induces the silencing of NbAGO1-1H and PDS transcripts. The analysis of viral genomic RNA accumulation is shown in total RNA from infected tissues at 6 dpi. C, Semiquantitative RT-PCR analysis of the viral RNA (TBSV row), the PDS transcripts (PDS row), and the endogenous control gene CYCLOPHYLIN (CPH; CPH row). Double asterisks highlight the amplified products containing the indel. D and E, Visual inspection of the leaf whitening associated with PDS silencing in TBSV:indelH/PDS- and TBSV:PDS-infected plants, respectively. F, qRT-PCR of NbAGO1-1H and NbAGO1-1L transcripts in mock-inoculated and TBSV:indelH/PDS- and TBSV:PDS-infected plants. Data are presented as means ± se of three replicates; different letters denote significant differences at P ≤ 0.05.
The fragment that was used in the TBSV:NbAGO1-1 construct is homologous to both NbAGO1-1H and NbAGO1-1L sequences; accordingly, it induced silencing of both transcripts (see “Materials and Methods”; Supplemental Fig. S1). In order to specifically dissect the relevance of the two homeologs, we next developed a TBSV vector that carried the sequence of the indel H that should consequently silence solely NbAGO1-1H. As explained, NbAGO1-1H transcripts accumulate at higher levels in virus-infected plant tissues (Fig. 4A). Hence, the TBSV-mediated VIGS was expected to reveal the specific function of NbAGO1-1H in antiviral activity and/or symptom development during a viral infection. In the subsequently performed experiment, we applied a construct that carried an additional flanking sequence that was believed to also silence the N. benthamiana endogenous phytoene desaturase (PDS; TBSV:indelH/PDS in Fig. 7B, top). PDS is an enzyme that is involved in intermediate steps in carotene biosynthesis. Accordingly, virus-induced silencing of this gene was expected to result in decoloration of infected tissues, which, in turn, should enable us to visually follow the silencing capacity of the viral vector (Pignatta et al., 2007). As a positive control, a TBSV construct was used that silenced solely the PDS. Thus, N. benthamiana plants were inoculated with TBSV:PDS and TBSV:indelH/PDS in vitro transcripts, and the accumulation of viral RNA was monitored on systemic leaves. As shown in Figure 7B, viral genomic RNA was detectable; it reached a peak at 6 dpi and was comparable in both cases (Figure 7B, bottom, lanes 2 and 3). Semiquantitative reverse transcription (RT)-PCR on total RNA that was extracted from six apical leaves at the recovery stage further supported that the levels of TBSV:PDS and TBSV:indelH/PDS accumulation were comparable, since the amplification product appeared at the 21st cycle with both infective constructs (Fig. 7C). Note that the amplified product of the semiquantitative RT-PCR contained the indel H fragment, which, accordingly, was verified to be retained by the TBSV vector during plant infection and spread (Fig. 7C, double asterisks; Sanger sequence not shown). These results revealed that the two vectors could accumulate at comparable levels in systemic leaves; the indel H insert was ensured to neither alter the capability of the viral vector to move long distances nor to replicate and to accumulate in infected tissues.
When we estimated the levels of PDS transcripts in TBSV:PDS- and TBSV:indelH/PDS-infected plants, as expected, both constructs were found to reduce the PDS mRNA to a level that was at least 1,000 times lower than in mock-inoculated plants. That is, in mock-inoculated plants, a discrete amplified product appeared at cycle 15 in the semiquantitative RT-PCR, while it appeared at cycle 28 in plants that had been infected with each of the TBSV constructs (Fig. 7C). Unexpectedly, though, in the recovered leaves, the whitening symptoms, which were attributed to PDS silencing, turned out to be visually different when we compared the plants that were infected with the two constructs. Indeed, in the case of TBSV:indelH/PDS, the whitening appeared brighter and more widely distributed among all newly recovered leaves, and this phenotype was found in all biological tests (Fig. 7, D and E). Conversely, with TBSV:PDS, the whitening appeared noticeably more confined to the leaf veins, as shown in the top right images in Figure 7, D and E. When we measured the level of the NbAGO1-1L and NbAGO1-1H transcripts, TBSV:indelH/PDS was confirmed to be effective in the silencing of NbAGO1-1H. This became most obvious at 6 dpi, when the transcript was almost undetectable (Fig. 7F). In contrast, in the case of the TBSV:PDS-infected tissues, the NbAGO1-1H transcript accumulated at a higher level than the NbAGO1-1L mRNA (Fig. 7F), which recapitulated the situation of the CymRSV infection shown in Figure 2C. Therefore, we concluded that the NbAGO1-1H homeolog, besides acting as an antiviral component, also has a significant impact on plant symptom development (see below).
DISCUSSION
NbAGO1-1H and NbAGO1-1L Loci and Transcripts
Like the majority of flowering plants, N. benthamiana (n = 19) possesses an allopolyploid genome (Goodin et al., 2008), as it underwent a process of whole-genome duplication through the recombination of two or more genomes from yet unidentified Nicotiana spp. progenitors (alloploidization). In this study, we show that N. benthamiana possesses two NbAGO1-1 loci. Likely, the NbAGO1-1L and NbAGO1-1H loci derived from such genome duplications; therefore, they can be considered as AGO1 homeologs. Genome duplication, except in cases of redundancy, is often followed by changes in gene expression/gene specialization (subfunctionalization), evolution of a novel function (neofunctionalization), or gene loss (nonfunctionalization including the generation of pseudogenes; Comai et al., 2000). For example, a phylogenetic approach carried out on N. tabacum (a close relative of N. benthamiana) and its progenitors Nicotiana tomentosiformis and Nicotiana sylvestris showed that about 90% of all homeolog sequences (6% of all genes) maintained the expression of only one homeolog, since the second homeolog was lost after genome duplication and evolution (Bombarely et al., 2012a).
Instead, in the N. benthamiana allopolyploid context, we show that both NbAGO1-1L and NbAGO1-1H homeologs retained the full capacity to transcribe mRNAs. Strikingly, a previous study (Nakasugi et al., 2013), which aimed to identify all the N. benthamiana RNA-silencing genes, did not highlight the presence of the NbAGO1-1L transcript. The authors describe two AGO1 transcripts, named AGO1a and AGO1b, both containing the indel. That the NbAGO1-1L transcript was missed is probably due to the fact that the NbAGO1 transcript that lacks the 18-nucleotide-long indel was poorly represented in the unigene data set (i.e. one out of eight AGO1.1 species; see above and Supplemental Fig. S3). Another explanation may be that the 18-nucleotide-long insertion in the AGO1 transcript is conserved among the Solanaceae (Fig. 2A, top) and, accordingly, was used as a reference sequence in the RNA sequencing assembly processes.
Our findings indicate that an 18-nucleotide-long GC-rich element in the NbAGO1-1 mRNA has important functional implications for the modulation of AGO1 expression in N. benthamiana. Interestingly, the element is missing in only one type of the known NbAGO1-1 mRNAs (Fig. 2A), and it has no evident consequences, either on the expression or on the functionality of the proteins, at least in the in vitro functional analysis performed here (Fig. 6; see below). Hence, it may be hypothesized that the speciation of N. benthamiana following allopolyploidization has resulted in two alternative transcripts of AGO1-1, the expression of which is differently regulated by miRNA-mediated cleavage at the posttranscriptional level. One interpretation of our findings involves the possibility that the NbAGO1-1H locus developed from the NbAGO1-1L locus and that this led to the expression of an mRNA that is less accessible to miR168 cleavage.
Regarding the level of expression of the two transcripts, our qRT-PCR data indicate that, in naive plants, the H form of NbAGO1-1 is slightly less abundant than the L form (i.e. 1:2; Figs. 4A and 7F, mock-inoculated plants). This could be due to a different transcriptional control, which deserves further specific investigation. It also remains unclear whether the two transcripts coexist in the same cells and whether they are spatially and/or temporally separated from the regulatory RISC (Souret et al., 2004; Brodersen et al., 2008).
The Indel and the miR168-Mediated Regulation of Transcripts
In a previous study on the activity of human RISC, Ameres et al. (2007) revealed that the accessibility of the target site directly correlates with the efficiency of the cleavage reaction. The two NbAGO1-1 homeolog transcripts differ in the 18-nucleotide-long indel, which is located only eight nucleotides upstream of the miR168 target site (Fig. 1C), and our prediction as well as experimental data suggest that this has considerable consequences for the formation of the secondary RNA structure of the miR168 target site. The presence of the indel in NbAGO1-1H appears to mask the miR168 target site and, thus, may render miR168 posttranscriptional regulation more difficult. This is in contrast with the situation with NbAGO1-1L, which was indicated to be clearly more accessible to the miR168 seed sequence (Fig. 3, A and B). This notion was further supported when we transferred the same sequence stretch into the context of the GFP reporter construct. The 18-nucleotide-long insertion had an evident effect on protein expression and mRNA accumulation, likely through a different sensitivity to miR168-mediated PTGS (Fig. 3, C and D). The idea that miR168-mediated cleavage of NbAGO1-1H occurs less efficiently (or less frequently) than NbAGO1-1L cleavage is also fueled by the 3′- and 5′-RACE data that were obtained in the GFP reporter gene context and during the in vivo analysis (Figs. 2B and 3C). Our analyses were performed with only one stretch of the NbAGO1-1H or NbAGO1-1L transcript, and we are aware that the results may not necessarily reproduce the same functionality in the whole RNA context. Unfortunately, a further detailed analysis in planta is impeded by the close similarity of the two homeologs. However, the sum of in silico, in vitro, and in vivo data supports our model, proposing that the 18-nucleotide-long indel represents an important modulator of the RNA structure that involves also the miR168 target site, which, when it is present, may confer a lower sensitivity, and upon its loss, may confer a higher sensitivity, to miR168-guided cleavage.
These data prompted us to look at transcript data that are available for other Solanaceae spp. and to search for variants that may have a similar impact on the miR168 target sites. Interestingly, the Solanum lycopersicum AGO1A-like and AGO1B (and Solanum tuberosum AGO1-like and AGO1B-like) alignment reveals one 21-nucleotide-long indel that is located 22 nucleotides downstream of the miR168 target site (Supplemental Fig. S6). In silico folding of these regions suggests that this indel may affect the secondary structure and accessibility of the miR168 target site of the Solanum spp. AGO1 mRNA in a similar way to that found here with NbAGO1-1H and NbAGO1-1L of N. benthamiana (data not shown).
Moreover, a recent S. lycopersicum genome-wide analysis of 5′-uncapped mRNAs (i.e. products of miRNA cleavages; German et al., 2008) suggests that both S. lycopersicum AGO1A-like and AGO1B are controlled by miR168 (Lopez-Gomollon et al., 2012). This is an additional indication that the presence of indels around the miR168 target site does not abolish cleavage-based posttranscriptional regulation events. Instead, these elements appear to play a role in the fine-tuning of the expression of duplicate genes and homeologs during conditions of miR168 up-regulation, such as under stress induced by viral infections.
NbAGO1-1 Homeologs in Viral Infection
N. benthamiana is widely considered as a model host for virus-plant interaction studies, and it has also been successfully used in molecular studies of RNA silencing-based plant-virus interactions with tombusvirus systems. Here, we applied CymRSV because the functionality of miR168 in N. benthamiana during a CymRSV infection was convincingly demonstrated by Várallyay et al. (2014). Indeed, we considered this virus system most adequate to dissect the miR168-directed regulation of the NbAGO1-1 mRNAs for the following reasons: (1) N. benthamiana plants show a strong accumulation of miR168 associated with a CymRSV infection (Fig. 4B; Várallyay et al., 2010); (2) while not binding miR168 (Várallyay et al., 2014), the viral suppressor P19 is supposed to effectively sequester vsiRNAs and to impede their interference with endogenous and physiological AGO1 functionalities (at least during the initial stages of infection and spread); and (3) the level of NbAGO1-1 mRNA seems to increase in CymRSV-infected plants, but this is not associated with an accumulation of mRNA cleavage products (Várallyay et al., 2010). Earlier works detected miRNA-mediated NbAGO1 mRNA cleavage in virus-infected cells at a lower extent than in naive cells (Várallyay et al., 2010), and our findings are consistent with these data, if we consider solely the situation with NbAGO1-1H (Fig. 2C). Várallyay et al. (2010) also observed a reduction of AGO1 protein accumulation and attributed this contradiction (i.e. up-regulation of AGO1 mRNA and miR168 but no increase of 5′ cleavage products) to an miRNA-mediated translation inhibition of the mRNA. Obviously, the above studies and conclusions did not discriminate between the two homeolog transcripts and were based on measurements of the total amount of both NbAGO1-1 mRNAs. As explained above, the two NbAGO1-1 homeologs were targeted and cleaved by miR168; however, the cleavage remnants of the H homeolog were less detectable by RACE analysis. Thus, it is conceivable that the measured cleavage products mainly derived from the L version of the NbAGO1 mRNA, while the H version, which is cleaved to a considerably lower extent, indeed is translationally repressed, as suggested by Várallyay et al. (2010). Future experiments need to address this interesting aspect.
A VIGS strategy was used as a preliminary approach to dissect the functionality of the two NbAGO1-1 homeologs. This strategy was based on the widely accepted assumption that AGO1 represents a major effector of PTGS of endogenous genes (Bologna and Voinnet, 2014). Thus, plants that were infected with TBSV:NbAGO1-1, which silenced both NbAGO1-1 homeologs, underwent a quick necrotic decline (Fig. 7A). Considering that both NbAGO1-1H and NbAGO1-1L are able to incorporate siRNAs and to reconstitute RISC active in cleaving viral RNA in vitro (Fig. 6), these data suggest that both homeolog genes together constitute a solid layer of defense against tombusvirus infections. In other words, both homeologs were indicated to be determinants of the susceptibility of N. benthamiana to infections with tombusviruses.
The application of a construct that was able to silence the endogenous PDS gene and specifically NbAGO1-1H highlighted another intriguing aspect. While at 6 dpi, we could achieve an efficient silencing of NbAGO1-1H (Fig. 7F), no differences in viral accumulation or necrosis were observed in comparison with the control (Fig. 7, B–E). However, as explained in “Results,” the whitening symptoms, which were associated with the silencing of the endogenous PDS gene, revealed evident differences between plants that were treated with TBSV:indelH/PDS and the TBSV:PDS control plants (Fig. 7, D and E). Closely consistent with our earlier data, we found the NbAGO1-1L transcript to be expressed almost twice as much as NbAGO1-1H in the mock-inoculated plants (Figs. 4A and 7F). During the infection, this ratio turned around (i.e. the amount of the NbAGO1-1L transcript decreased and the amount of NbAGO1-1H increased), which was explained again by the increase of miR168 expression and the lower sensitivity of NbAGO1-1H to miR168 cleavage. When forcing the silencing of NbAGO1-1H using VIGS, the plants showed again an increasing amount of the L form (Fig. 7F). This may suggest that the loss of the H form is somehow compensated by a higher gene expression of the L form to control the invasion of the virus.
The current status of our findings clearly supports the view that the presence of two forms of AGO1-1 represents an evolutionary advantage for the plant. Thus, both AGO1-1 forms were indicated to operate in concert to ensure basic mRNA regulatory control during conditions of stress, such as a viral infection where massive amounts of vsiRNAs are produced and where miR168 expression is up-regulated. Along this line, the presence of short indels in close proximity to miRNA target sites may represent a new type of genetic strategy to fine-tune the miRNA-mediated posttranscriptional regulation of genes under stress.
This speculation is, at least in part, supported by recent reports with Arabidopsis showing (1) the necessary presence of two specialized pools of AGO1 that are loaded with single classes of short RNA duplexes (i.e. with either miRNAs or viral siRNAs; Schott et al., 2012) and (2) the evidence that recovery and VIGS are mediated by different host factors, including AGO1 (Ma et al., 2015). Thus, our data add another layer of regulation of the AGO1 homeostasis in plants under external stimuli. Our findings are in line with an earlier report of Li et al. (2012), who observed that miR168 overexpression does not necessarily correlate with lower AGO1 mRNA levels.
As such, this study provides an important basis for future molecular, biochemical, and genome-wide studies aimed at unraveling the mechanisms of the functional diversification of genes involved in RNA silencing, with a particular relevance in the context of polyploidy.
MATERIALS AND METHODS
Bioinformatics Analysis
The draft genome of Nicotiana benthamiana (version 0.4.4) was downloaded from http://solgenomics.net/organism/Nicotiana_benthamiana/genome (Bombarely et al., 2012b). It comprises a set of 140,890 scaffolds (with a total length of 2,593,640,036 bp), which were imported into the CLC Genomics Workbench software (version 5.5) for further analysis. The scaffolds were screened for the presence of NbAGO1 (National Center for Biotechnology Information accession number DQ321488; 3,156 bp). Sequence matches were found on two distinct scaffolds, SCF00034990 and SCF00010009. From a pairwise alignment between the two alternative NbAGO1 gene sequences, point mutations (single-nucleotide and deletion-insertion polymorphisms) were detected as well as an 18-bp deletion (on scaffold SCF00034990). We defined the transcript containing the full gene sequence as NbAGO1-1H; the variant containing the 18-bp deletion was termed NbAGO1-1L. The intron/exon structure was determined through analysis with GENSCAN (http://genes.mit.edu/GENSCANinfo.html; Burge and Karlin, 1997) accompanied by manual interpretation.
To retrieve all the annotated and putative AGO1-like gene sequences available in databases, the NbAGO1-1H protein sequence was submitted to tBLASTn using the plant taxid database. Annotated or predicted AGO1-like mRNAs containing putative miR168 target sequences were selected and multialigned with ClustalW (Thompson et al., 1994; DNA weight matrix = default value, gap open = 100, and gap extension = 10).
cDNA of NbAGO1-1 Transcripts, GFP Sensor, and Agroinfiltration
Total RNA was extracted from 100 mg of expanded leaf tissues from N. benthamiana plants using TriReagent (Sigma) following the manual’s instructions. Upon DNaseI (Ambion) treatment to ensure the elimination of traces of DNA, the polyadenylated fraction was enriched using the Ambion Poly(A)Purist Kit (Life Technologies). The polyadenylated fraction was used for randomized cDNA generation. Oligonucleotides NbAGOmiRNAS and NbAGOmiRNAAS (Várallyay et al., 2010) were used for PCR amplification of the approximately 120-bp-long fragment containing the miR168 target site. Two PCR species (data not shown), almost indistinguishable in size, were observed on agarose gels. Therefore, the PCR was first loaded and separated through a 2% (w/v) agarose gel and excised from the very top and very bottom of the doublet. Then, the two amplified species were again separated via an 8% (w/v) polyacrylamide gel. From the latter gel, the species were cloned into the pGEM T-easy vector (Promega) and sequenced.
PCR product species of target sequences H and L were placed into the SmaI-linearized plasmid pAJ as described previously (Pantaleo et al., 2007). Sanger sequencing was used to verify the orientation of the cloned fragments. Sensor constructs pAJ GFP NbAGO1-1L (256-379) and pAJ-GFP NbAGO1-1H (256-397) were transferred into the Agrobacterium tumefaciens strain C58C1 as described previously (Pantaleo and Burgyán, 2008) and then agroinfiltrated into well-expanded 6- to 10-leaf-old N. benthamiana. After 3 dpi, the leaves were analyzed under the UV lamp.
5′- and 3′-RACE
5′-RACE analysis of miR168-cleaved NbAGO1-1 was performed as described previously (Shimura et al., 2011) with the polyadenylated fraction obtained from total RNA of N. benthamiana. Polyadenylated RNA was obtained with the Ambion Poly(A)Purist Kit (Life Technologies) following the manual’s instructions. A gene-specific reverse oligonucleotide for PCR having the sequence 5′-CTTCCATATGGTACAGGCTGA-3′ was designed downstream of the expected miR168 cleavage site.
3′-RACE of the GFP sensor sequence GFP-NbAGO1-1 was carried out on the RNA extracted from the agroinfiltrated spots as described previously (Pantaleo et al., 2007). After RACE PCR amplification, the gel slice corresponding to 150 to 200 nucleotides in size was excised, and the DNA was eluted, cloned, and Sanger sequenced.
CymRSV Infection and NbAGO1-1 mRNA qRT-PCR
The plasmid encoding the CymRSV RNA (Burgyan et al., 1990) was linearized with SmaI, ethanol precipitated, and transcribed in vitro using T7 RNA polymerase. N. benthamiana plants were inoculated with the viral RNA transcript as described (Rubino et al., 1992). Total RNA was extracted from leaf tissues with TriReagent (Invitrogen) following the manufacturer’s instructions. We analyzed leaves mock inoculated and at 3, 4, 6, and 10 dpi. Primers for qRT-PCR were designed taking advantage of the 18-nucleotide-long indel, in order to discriminate the homeologs NbAGO1-1H and NbAGO1-1L: NbAGO1-1H for, 5′-GCCATGGGGCACCTTCTG-3′; NbAGO1-1H rev, 5′-GAGACGAGGAACCAGCCTC-3′; NbAGO1-1L for, 5′-ATCAACGAGGTGGAGGACAA-3′; and NbAGO1-1L rev, 5′-GGTACTGGTGGCCGTGC-3′. Primers for qRT-PCR amplification of GFP mRNA were GFP for (5′-CGATGGCCCTGTCCTTTTAC-3′) and GFP rev (5′-GGTCTCTCTTTTCGTTGGGATCT-3′). First strand cDNA synthesis was performed using 5 μg of total RNA treated with DNase and the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Relative expression was calculated based on the comparative cycle threshold method as described by Livak and Schmittgen (2001). The PCR mix (10 μL) contained 5 μL of PowerSYBR Green master mix (Life Technologies), 0.25 μm of each primer, and 1 μL of cDNA diluted 1:10. Cycling conditions for NbAGO1-1H and NbAGO1-1L primer pairs consisted of initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, and 65°C for 1 min. Cycling conditions for GFP amplification consisted of initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min. Specific annealing of the primers was controlled on dissociation kinetics performed at the end of each PCR run. Expression of the CPH gene was used as the normalization factor in all samples as reported (Havelda et al., 2008), since CPH concentration in N. benthamiana is constant during virus infection. Transcript level was expressed as the mean and se calculated for three replicates. Data were statistically analyzed using the ANOVA F test (P ≤ 0.05).
Cell Culture and Preparation of Cytoplasmic BY-2 Cell Extract
Nicotiana tabacum BY-2 cells were cultured as described (Gursinsky et al., 2009) at 23°C in Murashige and Skoog liquid medium. Evacuolated BY-2 protoplasts to prepare cytoplasmic extract (BYL) were obtained by Percoll gradient centrifugation (Komoda et al., 2007; Gursinsky et al., 2009).
In Vitro Transcription
To generate plasmids for the production of NbAGO1-1H and NbAGO1-1L mRNAs, the corresponding open reading frames were amplified by PCR and inserted between the XbaI and SmaI sites of a modified pSP64-poly(A) vector (Promega) that contained an additional SwaI site downstream of the polyadenylated sequence.
Transcription and further treatment of the transcript were performed using standard procedures. Transcripts encoding firefly luciferase were generated by SP6 RNA polymerase (Thermo Scientific) from the XhoI-linearized plasmid pSP-luc(+) (Promega). AGO mRNAs were synthesized in the presence of the monomethylated cap analog m7GP3G (Jena Biosciences) from the SwaI-linearized plasmids using SP6 RNA polymerase. To generate the GFP target RNA, a 432-bp sequence was amplified by PCR from plasmid pGFP-C1 with the T7 promoter sequence included in the forward primer. Radioactive labeling was performed using standard conditions. TBSV defective interfering RNA was synthesized by T7 RNA polymerase from an SmaI-linearized template DNA (Schuck et al., 2013).
siRNAs
RNA oligonucleotides were purchased from Biomers. The sequences of the gf698 siRNA duplex were 5′-UAGUUCAUCCAUGCCAUGUGUA-3′ (guide strand) and 5′-CACAUGGCAUGGAUGAACUAUA-3′ (passenger strand). The sequences of the TBSV-derived vsiRNAs were 5′-UAUCCGACCAUAGGCCCAUGU-3′ (guide strand) and 5′-AUGGGCCUAUGGUCGGAUAAG-3′ (passenger strand). To produce siRNA duplexes, the single-stranded RNAs were incubated in annealing buffer (30 mm HEPES-KOH, pH 7.4, 100 mm potassium acetate, and 2 mm magnesium acetate) for 1 min at 90°C and annealed for 60 min at 37°C. All siRNAs used were nonphosphorylated.
In Vitro Slicer Assay
To generate AGO1 variants and siRNA-programmed AGO/RISC in vitro, the AGO mRNAs were in vitro translated in 50% (v/v) BYL in the previously described conditions (Gursinsky et al., 2009). Briefly, 1.5 µg of AGO mRNA was translated in a 20-µL reaction in the presence of 50 nm synthetic siRNA for 60 min. To measure slicer activity, the same amount of siRNA was added again, and the reaction was continued for another 90 min. Two micrograms of firefly luciferase (competitor) mRNA and the 32P-labeled target RNA (50 fmol) were added, and the cleavage reaction was performed for another 15 min. Total RNA was isolated from the reaction by treatment with 20 µg of Proteinase K in the presence of 0.5% (w/v) SDS for 30 min at 37°C, followed by extraction with 1 volume of chloroform and ethanol precipitation. 32P-labeled products were separated on 5% (w/v) Tris-borate polyacrylamide gels containing 8 m urea and visualized by phosphor imaging.
Virus-Induced RNA Silencing
The TBSV viral vector (pPD-A4) was kindly provided by Dr. M. Turina. The vector TBSV:NbAGO1-1 was obtained by exchanging the PDS fragment in pPD-A4 with an XhoI (Fermentas)-digested fragment obtained by PCR amplification of the NbAGO1-1 gene (positions 2,268–2,351; Supplemental Fig. S1) using 5′-TGCTCGAGCATACCCATGTGGCCTTGTCTTCAAG-3′ and 5′-ACACTCGAGAATATCACGTTCTCTC-3′ (XhoI restriction sites underlined) as oligonucleotides and total DNA extracted from leaves (Rubino et al., 1992) as substrate. For TBSV:indelH/PDS, oligonucleotides pdsXhoIrev (5′-AGGACACTCGAGCAGGAGGGTTACC-3′) and TBSVAGO11Hfor (5′-TAGTGCTCGAGGCACCTTCTGGTGGCCCTCCTCGGCACGAGCTTTCGATG-3′; forward oligonucleotides in boldface) were used to obtain the fragment of N. benthamiana PDS (positions 878–973 of the ref_seq DQ469932) with the flanking 25-nucleotide-long sequence of the NbAGO1-1H indel. All constructs were SmaI restricted, and infectious RNAs were generated and inoculated as described previously (Rubino et al., 1992).
Semiquantitative RT-PCR Analysis
To control equal cDNA amounts in each reaction, PCR was performed with primers corresponding to CPH (see above). Primer sequences were as follows: TBSV Forward, 5′-GAGGGTTACCATCTAAAAAGGCC-3′; TBSV Reverse, 5′-CTTGTTCGTATTCAGTATCC-3′; PDS Forward, 5′-GATGCWACRATGAAGGAACTAGC-3′; PDS Reverse, 5′-GCCGACARGGTTCACAACCTG-3′.
Chemical Probing of RNA Secondary Structure and Primer Extension Analysis
Evaluation of the RNA secondary structure was done using the protocol released by Yu et al. (1999) with minor modifications. The modifying agent used was DMS (Merck). DMS modification was performed with 1 µg of in vitro-transcribed RNA and 32 mm DMS in a 200-µL reaction mixture containing 20 mm HEPES-KOH (pH 7.9), 60 mm KCl, and 12 mm MgCl2 at 30°C for 5 min. The reaction was stopped by the addition of an equal volume of DMS stop buffer containing 0.6 m sodium acetate, 0.4 m β-mercaptoethanol, 0.4 m Tris-HCl (pH 7.5), and 10 mm EDTA. The modified RNA was then ethanol precipitated with 10 µg of tRNA as a carrier.
Chemically modified RNA molecules were mixed with 1 pmol of 5′ end-labeled DNA primer (5′-CGCACGCGTGGTACAGGCTGA-3′). Primer extension was performed at 42°C for 60 min with 100 units of RevertAid Reverse Transcriptase (Thermo Scientific) at standard conditions as recommended by the manufacturer. The locations of modification sites were determined by PAGE of the primer extension products (see above). In parallel, the corresponding DNA sequences were determined by dideoxynucleotide sequencing (DNA cycle sequencing kit; Jena Bioscience) with the same 5′ end-labeled primer.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KR942296 and KR942297.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence alignment of NbAGO1-1L and NbAGO1-1H loci.
Supplemental Figure S2. PCR-based strategy for validating NbAGO1-1L and NbAGO1-1H loci.
Supplemental Figure S3. List of NbAGO1-1H and NbAGO1-1L transcript species found in the RNA-Seq data set.
Supplemental Figure S4. Multiple alignment of miR168 family members.
Supplemental Figure S5. qRT-PCR for discriminating NbAGO1-1H and NbAGO1-1L.
Supplemental Figure S6. Multiple alignment of annotated ARGONAUTE1 mRNAs of Solanum lycopersicum and Solanum tuberosum.
Supplementary Material
Acknowledgments
We thank David S. Horner for a critical review of the article.
Glossary
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- PTGS
posttranscriptional gene silencing
- vsiRNA
virus-derived small interfering RNA
- CymRSV
Cymbidium ringspot virus
- TBSV
Tomato bushy stunt virus
- siRNA
small interfering RNA
- indel
insertion/deletion
- VIGS
virus-induced gene silencing
- cDNA
complementary DNA
- DMS
dimethyl sulfate
- dpa
days post agroinfiltration
- qRT
quantitative reverse transcription
- dpi
days post inoculation
- BYL
BY-2 cell lysate
- RT
reverse transcription
Footnotes
This work was supported by the Italian Ministry of Economy and Finance through the Consiglio Nazionale delle Ricerche (CNR; grant no. C.I.S.I.A. Legge 191/2009 and by the AQUA project), by the Dipartimento Agro-Alimentare-CNR (to V.P.), by a CNR Short Term Mobility Grant 2014 (to V.P.), and by the Deutsche Forschungsgemeinschaft (grant no. BE1885/7–2, project B6 of the Research Training Group 1591, and project A7 of the Collaborative Research Center 648 to T.G., S.F., and S.-E.B.) at Martin Luther University Halle-Wittenberg.
Articles can be viewed without a subscription.
References
- Ameres SL, Martinez J, Schroeder R (2007) Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65: 473–503 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Edwards KD, Sanchez-Tamburrino J, Mueller LA (2012a) Deciphering the complex leaf transcriptome of the allotetraploid species Nicotiana tabacum: a phylogenomic perspective. BMC Genomics 13: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB (2012b) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25: 1523–1530 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Burgyán J, Havelda Z (2011) Viral suppressors of RNA silencing. Trends Plant Sci 16: 265–272 [DOI] [PubMed] [Google Scholar]
- Burgyan J, Nagy PD, Russo M (1990) Synthesis of infectious RNA from full-length cloned cDNA to RNA of cymbidium ringspot tombusvirus. J Gen Virol 71: 1857–1860 [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B (2000) Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Bovi A, Dalmay T, Burgyan J (2007) The p122 subunit of Tobacco mosaic virus replicase is a potent silencing suppressor and compromises both siRNA and miRNA mediated pathways. J Virol 81: 11768–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Pantaleo V, Burgyán J (2009) RNA silencing: an antiviral mechanism. Adv Virus Res 75: 35–71 [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Zaitlin D, Naidu RA, Lommel SA (2008) Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 21: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Gursinsky T, Schulz B, Behrens SE (2009) Replication of Tomato bushy stunt virus RNA in a plant in vitro system. Virology 390: 250–260 [DOI] [PubMed] [Google Scholar]
- Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstädt S, Carr JP, Baulcombe DC (2011) An antiviral defense role of AGO2 in plants. PLoS ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, Meister G (2013) Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat Struct Mol Biol 20: 814–817 [DOI] [PubMed] [Google Scholar]
- Havelda Z, Várallyay E, Válóczi A, Burgyán J (2008) Plant virus infection-induced persistent host gene downregulation in systemically infected leaves. Plant J 55: 278–288 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M (2010) In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y (2013) Molecular insights into microRNA-mediated translational repression in plants. Mol Cell 52: 591–601 [DOI] [PubMed] [Google Scholar]
- Jones L, Keining T, Eamens A, Vaistij FE (2006) Virus-induced gene silencing of Argonaute genes in Nicotiana benthamiana demonstrates that extensive systemic silencing requires Argonaute1-Like and Argonaute4-Like genes. Plant Physiol 141: 598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2005) The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev Biol 280: 504–517 [DOI] [PubMed] [Google Scholar]
- Komoda K, Mawatari N, Hagiwara-Komoda Y, Naito S, Ishikawa M (2007) Identification of a ribonucleoprotein intermediate of tomato mosaic virus RNA replication complex formation. J Virol 81: 2584–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J 25: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crété P, Voinnet O, Robaglia C (2009) Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Q, Jin C, Lai L, Feng J, Chen S, Chen J (2011) Tobacco microRNAs prediction and their expression infected with Cucumber mosaic virus and Potato virus X. Mol Biol Rep 38: 1523–1531 [DOI] [PubMed] [Google Scholar]
- Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, Jin H, Zhang D, Liang W (2012) Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol 158: 1279–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y (2007) Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14: 287–294 [DOI] [PubMed] [Google Scholar]
- Lopez-Gomollon S, Mohorianu I, Szittya G, Moulton V, Dalmay T (2012) Diverse correlation patterns between microRNAs and their targets during tomato fruit development indicates different modes of microRNA actions. Planta 236: 1875–1887 [DOI] [PubMed] [Google Scholar]
- Ma X, Nicole MC, Meteignier LV, Hong N, Wang G, Moffett P (2015) Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J Exp Bot 66: 919–932 [DOI] [PubMed] [Google Scholar]
- Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM (2013) De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS ONE 8: e59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro B, Russo M, Pantaleo V, Rubino L (2006) Cytological analysis of Saccharomyces cerevisiae cells supporting cymbidium ringspot virus defective interfering RNA replication. J Gen Virol 87: 705–714 [DOI] [PubMed] [Google Scholar]
- Omarov R, Sparks K, Smith L, Zindovic J, Scholthof HB (2006) Biological relevance of a stable biochemical interaction between the tombusvirus-encoded P19 and short interfering RNAs. J Virol 80: 3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T, Nagy PD (2003) Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314: 315–325 [DOI] [PubMed] [Google Scholar]
- Pantaleo V, Burgyán J (2008) Cymbidium ringspot virus harnesses RNA silencing to control the accumulation of virus parasite satellite RNA. J Virol 82: 11851–11858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo V, Rubino L, Russo M (2003) Replication of Carnation Italian ringspot virus defective interfering RNA in Saccharomyces cerevisiae. J Virol 77: 2116–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Burgyán J (2007) Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J Virol 81: 3797–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O (2004) In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 18: 2237–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Kumar P, Turina M, Dandekar A, Falk BW (2007) Quantitative analysis of efficient endogenous gene silencing in Nicotiana benthamiana plants using tomato bushy stunt virus vectors that retain the capsid protein gene. Mol Plant Microbe Interact 20: 609–618 [DOI] [PubMed] [Google Scholar]
- Qiu W, Park JW, Scholthof HB (2002) Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant Microbe Interact 15: 269–280 [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Rubino L, Carrington JC, Russo M (1992) Biologically active cymbidium ringspot virus satellite RNA in transgenic plants suppresses accumulation of DI RNA. Virology 188: 429–437 [DOI] [PubMed] [Google Scholar]
- Scholthof HB, Alvarado VY, Vega-Arreguin JC, Ciomperlik J, Odokonyero D, Brosseau C, Jaubert M, Zamora A, Moffett P (2011a) Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol 156: 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof KB, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, et al. (2011b) Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 12: 938–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott G, Mari-Ordonez A, Himber C, Alioua A, Voinnet O, Dunoyer P (2012) Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J 31: 2553–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck J, Gursinsky T, Pantaleo V, Burgyán J, Behrens SE (2013) AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res 41: 5090–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, Burgyán J, Masuta C (2011) A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, McKhann H, Garcion C, Vaucheret H, Sandberg G, et al. (2005) Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17: 1343–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Szittya G, Molnár A, Silhavy D, Hornyik C, Burgyán J (2002) Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14: 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Pantaleo V, Toth G, Rusholme Pilcher RL, Moulton V, Burgyan J, Dalmay T (2010) Structural and functional analysis of viral siRNAs. PLoS Pathog 6: e1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y (2008) The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol 49: 493–500 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay E, Havelda Z (2013) Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol Plant Pathol 14: 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay É, Oláh E, Havelda Z (2014) Independent parallel functions of p19 plant viral suppressor of RNA silencing required for effective suppressor activity. Nucleic Acids Res 42: 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay E, Válóczi A, Agyi A, Burgyán J, Havelda Z (2010) Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J 29: 3507–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyán J, Hall TM (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2009) AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE 4: e6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP (2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura Y, Scholthof HB (2005) Tomato bushy stunt virus: a resilient model system to study virus-plant interactions. Mol Plant Pathol 6: 491–502 [DOI] [PubMed] [Google Scholar]
- Yu H, Grassmann CW, Behrens SE (1999) Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J Virol 73: 3638–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.