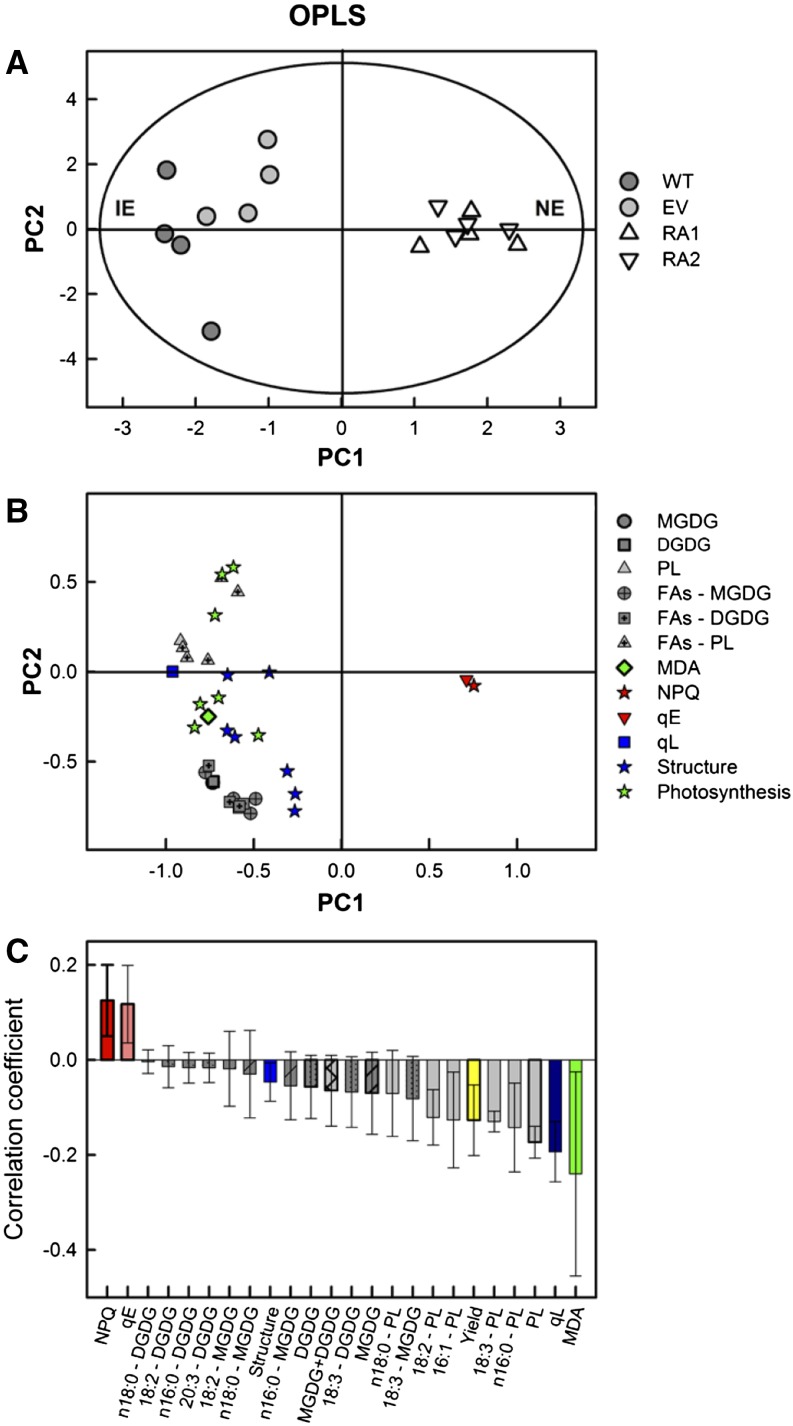
Figure 7.
Score (A), loading (B), and correlation coefficient plots (C) of orthogonal partial least squares (OPLS) of lipid classes, fatty acid composition, and MDA contents in isolated chloroplasts, chlorophyll fluorescence parameters measured in intact leaves (NPQ, ΦPSII, qE, and qL), and chloroplast proteins related to photosynthesis and proteins with structural activity. A, IE plants (wild type [WT] and empty vector [EV]), gray circles; NE plants (RA1 and RA2), white triangles. B, Each parameter is indicated with a different symbol: dark-gray circles, MGDG; dark-gray squares, DGDG; gray triangles, PL; dark-gray circles with a dot, MGDG fatty acids; dark-gray squares with a dot, DGDG fatty acids; gray triangles with a dot, PL fatty acids; green diamonds, MDA; red stars, NPQ; red down triangles, qE; blue squares, qL; blue stars, proteins with structural activity; and green stars, proteins related to photosynthesis. C, Parameter colors match those of the symbols in B. Only discriminant data with variable of importance for the projection (VIP) > 1 (for all except proteins) and VIP > 0.5 (proteins) are presented. Model fitness: Q2(Y) = 84%, R2(X) = 44%, r2 = 93%, and R2(Y) = 100% using one principal component; P = 0.00061, cross-validated ANOVA.