Figure 2. L1 activates the CLU gene promoter via an increase in STAT-1 expression.
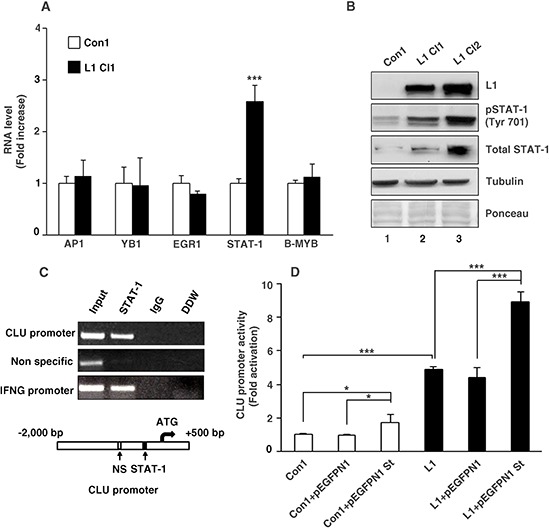
A. Quantitative RT-PCR analysis for RNA levels of transcription factors reported to regulate CLU transcription (AP-1, YB1, EGR1, STAT-1 and B-MYB) in L1 overexpressing (L1 Cl1) compared to empty vector-transfected (Con1) Ls174T cells. B. Western blot analysis for total STAT-1 and pSTAT-1 (Tyr 701) in Ls174T CRC cell clones stably transfected with L1 (L1 Cl1 and L1 Cl2) or with the control pcDNA3 plasmid (Con1). Tubulin antibody and total protein staining of the western blots with Ponceau were used for checking equal loading. C. Chromatin immunoprecipitation (Ch-IP)-based PCR analysis was employed using nuclear lysates from Ls174T cells stably transfected with L1 and primer sets that amplify the STAT-1-binding sequence, a negative control (Non specific) sequence and as positive control served a sequence from the interferon gamma (IFNG) promoter. As a control for the PCR analysis, a PCR reaction containing double-distilled water was used (DDW). DNA fragments conjugated with nuclear proteins were immunoprecipitated with mouse anti STAT-1 or non-immune goat IgG antibodies. Fragments containing the STAT-1-binding site and the control sites were amplified from the DNA obtained by Ch-IP. D. The CLU promoter reporter plasmid was co-transfected together with the pSV β-galactosidase control vector (for transfection efficiency normalization) into an Ls174T CRC cell control clone transfected with the empty plasmid (pcDNA3) (Con1), or an L1-overexpressing CRC cell clone. These cell clones were transiently transfected with the pEGFPN1 plasmid, or the STAT-1 expression plasmid pEGFPN1 St. Fold CLU promoter activation was determined after dividing luciferase activity by the values obtained with the empty reporter plasmid. *p < 0.05, ***p < 0.001. Error bars: ±S.D.
