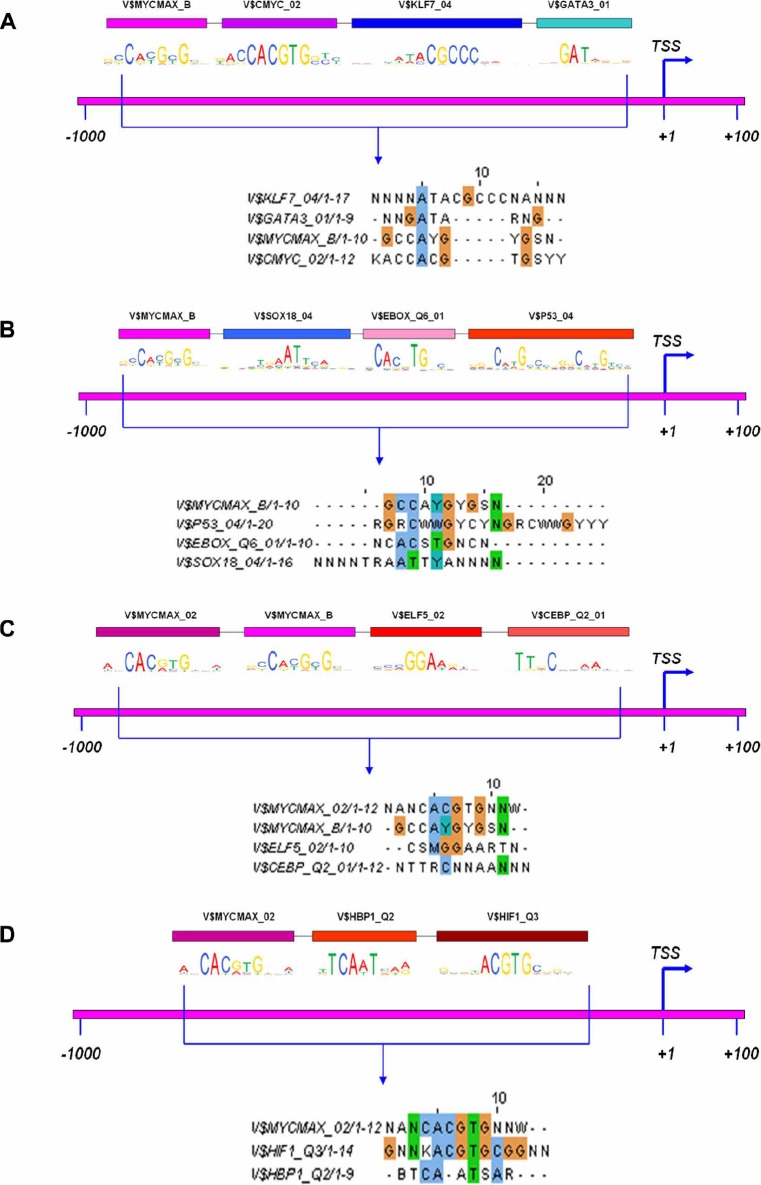
Figure 6. Functional transcription factor composite modules.
The co-occupancy of different transcription factor binding sites at gene specific promoters was analyzed. The promoter region with the best possible composite module is depicted with the consensus motif sequence locus. The software Clustal W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used for multiple sequence alignment (MSA) of motifs as to define overlapping regions. A. Composite module for down-regulated genes. 63, 67 and 96% had consensus binding sites for c-Myc, Klf7 and Gata3, respectively; the entire composite module fitted 46% of regulated genes. B. Alternate composite module for down-regulated genes. 92, 92 and 96% had consensus binding sites for c-Myc, Sox18 and P53, respectively; the entire composite module fitted 83% of regulated genes. C. Composite module for up-regulated tumor genes. 74, 96 and 93% had consensus binding sites for Myc, Elf5 and Cebpα, respectively; the entire composite module fitted 63% of up-regulated genes. D. Composite module for up-regulated non-tumor transgenic lung. 43, 25 and 53% genes had consensus binding sites for c-Myc, Hbp1 and Hif1, respectively.