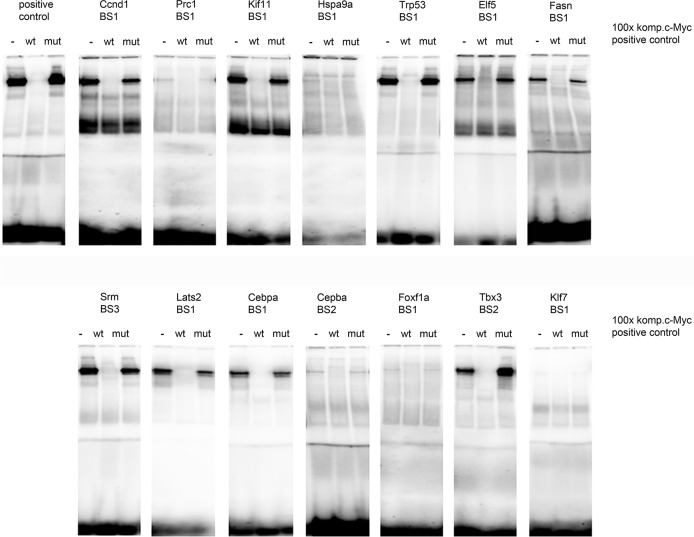
Figure 7. c-Myc DNA binding activity at gene specific promoters.
c-Myc DNA-binding at predicted gene-specific promoter sites of candidate genes was determined by EMSA. A total of 14 c-Myc binding sites were investigated using oligonucleotide probes described in Supplementary Table S8. Nuclear extracts from HeLa cells served as positive control (= positive control). DNA binding activity was assayed using probes specifically designed to recognize the predicted consensus binding site and is marked as – in the first lane of each gel. Specificity of DNA binding activity was determined in competition assays using 100-fold excess of the unlabeled probe (marked as wt = wild type) and by using a mutated (mut) probe whose nucleotide sequence was altered to be unable to recognize the core consensus binding site. Next to the positive control DNA binding activity was confirmed for Ccnd1, Prc1, Kif11, Trp53, Elf5, Fasn, Srm, Lats2, Cebpa, Foxf1 and Tbx3 and the observed bands were removed in competition assays with excess unlabelled probe (× 100 − fold) but not with the mutated probes. Notably, several commercially available C- and N- terminus directed antibodies were tested for their use in band shift assays but none of tested antibodies proved useful.