Figure 3. RNA-binding motif 4 [RBM4] enhances the skipping of neuronal polypyrimidine tract-binding protein [nPTB] exon 10 in colorectal cancer [CRC] cells.
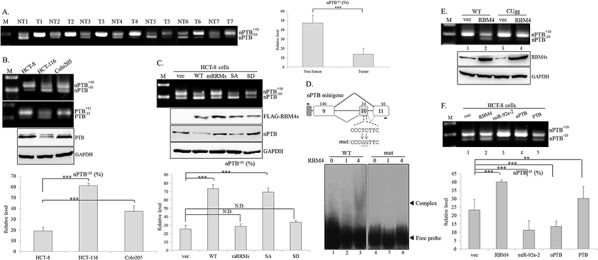
Total RNAs extracted from A. paired CRC tissues and B. CRC cell lines were analyzed by an RT-PCR with a specific primer set against the nPTB and PTB genes. Western blotting was performed with the indicated antibodies. C. Total RNAs were extracted from HCT-8 cells transfected with FLAG-tagged RBM4 and derived mutants. The splicing profile of nPTB and its protein expression were analyzed as previously described. The bar graph presents the relative level of nPTB−10 transcripts using TotalLab Quant Software [*p < 0.05; **p < 0.01; ***p < 0.005]. D. The diagram presents the sequence of the CU-element within nPTB exon 10. The mutant reporter contained pyrimidine-to-guanine nucleotide substitutions in the CU element. The mock eluate or recombinant His-tagged RBM4 protein was incubated with DIG-labeled probes. Mixtures were fractionated in 8% native acrylamide gels and transferred to Nylon membranes, followed by probing with an HRP-conjugated anti-DIG Fab fragment. E. The WT nPTB minigene or CUgg mutant was cotransfected with the empty vector or FLAG-RBM4 expression vector into HCT-8 cells. The PCR product of the spliced transcript was analyzed using electrophoresis on 2% agarose gels. F. The nPTB minigene was cotransfected with various expressing vectors into HCT-8 cells. The spliced transcript of the nPTB minigene was analyzed as described in the previous panel. The bar graph shows the relative level of nPTB−10 in three independent experiments using TotalLab Quant Software [*p < 0.05; **p < 0.01; ***p < 0.005].
