Figure 1. Sp1 transcriptionally regulates ZNF32 expression in response to oxidative stress.
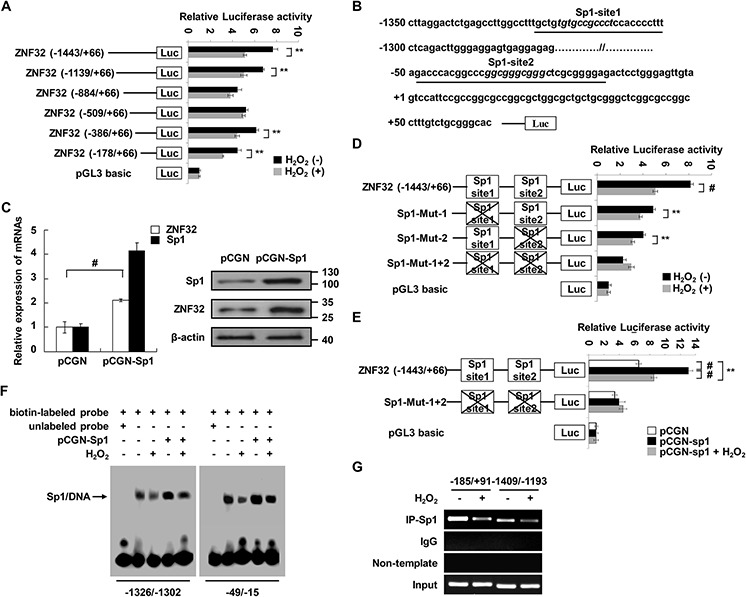
A. HEK293 cells were transiently transfected with ZNF32 promoter 5′-deletion mutant constructs, treated with 0.5 mM H2O2 for 24 h and then analyzed using a dual luciferase reporter assay. B. Schematic representation of the two Sp1-binding sites in the ZNF32 promoter. The consensus sequences for the predicted Sp1 sites (GC boxes) are shown in italics; the oligonucleotides used in EMSA are underlined; and the transcription start site is indicated by +1. C. HEK293 cells were transiently transfected with the indicated constructs and then subjected to qRT-PCR and immunoblot analysis. D. and E. HEK293 cells were transiently transfected with the indicated constructs, treated as in (A) and then analyzed using a dual luciferase reporter assay. F. Nuclear extracts from the indicated cells were incubated in biotin-labeled oligonucleotides corresponding to the ZNF32 promoter region (−1326/−1302 bp) or (−49/−15 bp). The arrow shows the specific DNA-protein complex. G. DNA fragments from HEK293 cells treated as in (A) were immunoprecipitated with Sp1-specific antibodies and analyzed via RT-PCR using the indicated primers. The data are presented as the mean values ± SEM. Each experiment was performed at least in triplicate, producing consistent results. *p < 0.05, **p < 0.01, #p < 0.001.
