Abstract
Staphylococcus xylosus is commonly used as starter culture for meat fermentation. Its technological properties are mainly characterized in vitro, but the molecular mechanisms for its adaptation to meat remain unknown. A global transcriptomic approach was used to determine these mechanisms. S. xylosus modulated the expression of about 40–50% of the total genes during its growth and survival in the meat model. The expression of many genes involved in DNA machinery and cell division, but also in cell lysis, was up-regulated. Considering that the S. xylosus population remained almost stable between 24 and 72 h of incubation, our results suggest a balance between cell division and cell lysis in the meat model. The expression of many genes encoding enzymes involved in glucose and lactate catabolism was up-regulated and revealed that glucose and lactate were used simultaneously. S. xylosus seemed to adapt to anaerobic conditions as revealed by the overexpression of two regulatory systems and several genes encoding cofactors required for respiration. In parallel, genes encoding transport of peptides and peptidases that could furnish amino acids were up-regulated and thus concomitantly a lot of genes involved in amino acid synthesis were down-regulated. Several genes involved in glutamate homeostasis were up-regulated. Finally, S. xylosus responded to the osmotic stress generated by salt added to the meat model by overexpressing genes involved in transport and synthesis of osmoprotectants, and Na+ and H+ extrusion.
Keywords: Staphylococcus xylosus, meat, transcriptome, nutrient adaptation, osmotic stress
Introduction
Meat fermentation is an old process that relies on indigenous microbiota and is nowadays carried out under controlled conditions with addition of starter cultures. Meat starter cultures are composed of a combination of lactic acid bacteria and coagulase-negative staphylococci (Ravyts et al., 2012). Staphylococcus xylosus and Staphylococcus carnosus are the two main staphylococcal species used as starter cultures. They are associated with different species of Lactobacillus, such as Lactobacillus sakei or Lactobacillus curvatus or Pediococcus. Studies over decades of microorganisms used as starters have yielded information about their potential functionalities in meat products. Lactic acid bacteria are well-known acidifiers and bacteriocin producers that affect the technological properties and the microbial stability of products (Leroy et al., 2006). Staphylococci are recognized to play an important role in the color formation through their nitrate reductase activity (Talon and Leroy, 2006). They also contribute to the flavor development of meat products via their catabolism of branched-chain amino acids in odorous volatile methyl compounds (Olesen and Stahnke, 2004; Tjener et al., 2004), their antioxidant properties avoiding rancidity (Barrière et al., 2001; Rosenstein et al., 2009) and their catabolism of pyruvate in diacetyl and acetoin responsible of buttery aroma (Søndergaard and Stahnke, 2002).
The development of -omic techniques has allowed the study of the metabolism of starter cultures in situ in the food matrix. The transcriptomic response of Lactococcus in milk or cheese has been established (Cretenet et al., 2011; Taïbi et al., 2011), while the proteome of Streptococcus thermophilus was studied in milk and the release of bacterial proteins was studied in cheese (Gagnaire et al., 2004; Derzelle et al., 2005).
Very few studies have focused on physiology of meat starter cultures in meat matrices. They concern essentially L. sakei. In vivo expression technology (IVET) was applied to investigate gene expression of L. sakei during meat fermentation (Hüfner et al., 2007). This study revealed the expression of genes encoding proteins that contribute to stress-related functions. A proteomic approach highlighted that L. sakei overexpressed dipeptidases in the presence of sarcoplasmic extract and proteins related to energy and pyrimidine metabolism in the presence of myofibrillar extract (Fadda et al., 2010). Concerning staphylococci, the single transcriptomic concerning the response of S. xylosus to nitrate and nitrite in meat revealed that S. xylosus was subject to nitrosative stress and overcame it by expressing genes involved in iron homeostasis and antioxidant defense (Vermassen et al., 2014).
Functional genomics give us an opportunity to gain insight into the physiological and metabolic capabilities of starters in situ in the food matrix. As the molecular mechanisms involved in the adaptation of the bacteria in meat products are little known, we analyzed the transcriptome of S. xylosus in a meat model incubated for 72 h at 22°C in conditions that mimic the fermentation step. The in situ response of S. xylosus was analyzed vs. the S. xylosus culture used as inoculum. The results highlighted a total change in gene expression during survival of S. xylosus in the meat model, which reflects adaptation to nutrients and mainly to osmotic stress.
Materials and methods
Preparation of the inoculum and inoculation of the meat model
The S. xylosus C2a strain is derived from the type strain DSM20267 cured of its endogenous plasmid pSX267. Its complete genome has been sequenced (LN554884). This strain grows and behaves in meat models and in batter as strains isolated from meat products (our unpublished data). We used this laboratory strain as it is the only one of this species with some genetic background. The strain was cultured overnight at 30°C with shaking (150 rpm) in a minimal medium as already described (Fiegler and Brückner, 1997). The culture was briefly centrifuged and part of the cell pellet called “the inoculum” was immediately frozen in liquid nitrogen before extraction of RNA. Another part of the cell pellet was resuspended in physiological serum and inoculated in a pork meat model containing 0.5% glucose and 0.47 M NaCl as already described in Vermassen et al. (2014). Petri dishes were completely filled with inoculated pork meat and incubated at 22°C in a wet atmosphere for up to 72 h. These conditions probably created a gradient of oxygen in the samples. At 24, 48, and 72 h of incubation at 22°C, 200 mg meat samples were taken and immediately frozen in liquid nitrogen to stabilize the bacterial RNA. Three independent experiments were done.
To evaluate the inoculum and the growth of S. xylosus in meat, bacteria were enumerated after serial dilution on plates of brain-heart infusion agar, which were incubated at 30°C for 24 h.
Physical and chemical analysis of the meat model
A digital pH meter (MP230, Mettler Toledo, Viroflay, France) with a Inlab® 413 electrode was used for measurement of meat pH.
Meat samples (2 g) were homogenized for 30 min with 8 mL of distilled water. The homogenate was then centrifuged for 20 min at 10,000 g at 4°C and the supernatant was filtered. The analyses were carried out as previously described (Nouaille et al., 2009). Briefly, glucose and fermentation products were analyzed by high-performance liquid chromatography (HPLC) using a Bio-Rad HPX87H column under the following conditions: a temperature of 48°C, 5 mM H2SO4 as the eluent at a flow rate of 0.5 mL min−1 and dual detection by refractometer and UV analyses. Free amino acids were analyzed after protein precipitation of the supernatant and derivatization. The derivatives were separated by HPLC on a Hypersil AA-ODS column at 40°C by a linear gradient of acetate buffer (pH 7.2) with triethylamine (0.018%), tetrahydrofurane (0.3%) and acetonitrile (60%). Diode array detectors were used at 338 and 262 nm for derivatives.
RNA extraction, labeling and microarray analyses and validation
The RNA extraction and labeling of S. xylosus either from the inoculum or directly in meat after 24, 48, or 72 h of incubation were carried out as described in Vermassen et al. (2014).
A complete description of the array developed for S. xylosus C2a is available at the NCBI Gene Expression Omnibus (GEO) database under platform accession number GPL19201.
Microarrays were analyzed as described in Vermassen et al. (2014). Significant differences in the probe set intensities between the two conditions were identified using a linear model with an empirical Bayes method using all information probes to moderate the standard errors of the estimated log-fold changes (Smyth, 2004). The probabilities were corrected by the Benjamini-Hochberg procedure in order to control the false-discovery rate (FDR) with a p-value cut-off of 0.05. All the probes with an FDR = 0.05 are considered to be differentially expressed. Finally, a gene was considered to be differentially expressed if at least 50% of the corresponding probes were differentially expressed and if the ratio of expression was above 2 or lower than 0.5.
Microarray data were validated as described in Vermassen et al. (2014). The targeted genes for qPCR and primer sequences are listed in Supplementary Table 1. The analyses were performed on the same samples of RNA as used for the microarray experiments. The relative fold change of gene expression, using measured tuf housekeeping gene expression, was determined by the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Microarray data accession number
The microarray samples and data have been deposited in the GEO database under accession number GSE69743.
Results and discussion
Growth and survival of S. xylosus in the meat model and transcriptome profile
S. xylosus was collected at the early stationary phase of growth in a minimal medium and was inoculated in the meat model at 7.7 log CFU/g. The growth of S. xylosus in the meat model was exponential until 24 h and reached 9.0 log CFU/g and remained almost at this population level until the end of the experiment (72 h).
The in situ S. xylosus response revealed a global change in gene expression during its survival in the meat model by comparison with the inoculum. There were 1337 (658 down- and 679 up-regulated), 1326 (659 down- and 667 up-regulated), and 1070 (540 down- and 530 up-regulated) genes differentially expressed at 24, 48, and 72 h, respectively (Supplementary Table 2). Notably, 838 genes were differentially expressed at the three times of incubation. This indicated that major transcriptional changes had occurred at 24 h and lasted throughout the incubation (72 h). These genes were classified into different functional categories: the most represented being information storage and processing, cellular processes and metabolism.
To validate the microarray analysis independently, the relative expression of 45 differentially expressed genes representing more than 5% of the common genes with significantly modified expression was measured by qPCR (Supplementary Table 1). The microarray and qPCR results for the tested genes were strongly correlated for the three times of incubation (24 h: r2 = 0.931 and slope = 1.235x, 48 h r2 = 0.918 and slope = 1.273x, 72 h r2 = 0.893 and slope = 1.394x) and the expected trend in the expression pattern was confirmed (Supplementary Figure 1).
The inoculum of S. xylosus was grown in a minimal medium with glucose as sole carbon source, NH4 as nitrogen source, 3 vitamins and trace elements. It then has to adapt its physiology to a pork meat model medium with an approximate composition (expressed as g/per 100 g) of proteins (15–22), lipids (1.5–4.0), minerals and trace amounts of carbohydrate (Toldrá, 2007) with glucose and NaCl added in our conditions. Furthermore, it has to adapt to different environmental conditions: pH variations (pH 5.9 in meat vs. 7.0), solid medium vs. liquid one.
Cell division and cell lysis
Even though S. xylosus began a plateau phase at 24 h, it remained metabolically active in the meat model until 72 h, as revealed by the modulation of 60 genes involved in information processing and storage and 36 genes involved in cellular processes (cell division, cell wall/membrane biogenesis) (Table 1, Supplementary Table 2).
Table 1.
Genes of Staphylococcus xylosus discussed in this study differentially expressed over time in meat.
| Gene ID | Gene name | Description | Mean ratio of expression | ||
|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | |||
| CELL DIVISION AND CELL LYSIS | |||||
| Replication, recombination, repair, and transcription | |||||
| SXYL_01165 | dnaE | DNA polymerase III subunit alpha | 2.6 | 3.5 | 2.6 |
| SXYL_01799 | DNA polymerase X family protein | 2.4 | 2.6 | 2.0 | |
| SXYL_01529-30 | parCE | DNA topoisomerase 4 subunit A, B | 5.0* | 3.8* | 2.9* |
| SXYL_00016 | dnaB | Replicative DNA helicase | 4.9 | 4.4 | 3.7 |
| SXYL_01219 | ruvB | Holliday junction ATP-dependent DNA helicase RuvB | 4.2 | 3.4 | 2.5 |
| SXYL_01659 | recG | ATP-dependent DNA helicase RecG | 3.8 | 4.0 | 2.6 |
| SXYL_02129 | recQ1 | ATP-dependent DNA helicase RecQ | 4.8 | 7.0 | 5.0 |
| SXYL_01954 | addA | ATP-dependent helicase/nuclease subunit A | 4.0 | 3.4 | 2.4 |
| SXYL_00005-06 | gyrAB | DNA gyrase subunits A and B | 3.7* | 3.9* | 2.8* |
| SXYL_02427 | mfd | Transcription-repair-coupling factor | 5.8 | 6.9 | 4.2 |
| SXYL_01583 | mutS1 | DNA mismatch repair protein MutS | 3.6 | 4.2 | 2.9 |
| SXYL_01798 | mutS2 | Endonuclease MutS2 | 3.2 | 4.8 | 3.5 |
| SXYL_02373 | fusA | Elongation factor G | 3.5 | 2.4 | |
| SXYL_01273 | lepA | Elongation factor 4 | 8.4 | 8.2 | 5.9 |
| SXYL_00727 | infA | Translation initiation factor IF-1 | 5.8 | 3.2 | 2.5 |
| SXYL_01186 | infC | Translation initiation factor IF-3 | 6.2 | 4.1 | 3.9 |
| Nucleotides transport and metabolism | |||||
| SXYL_02522-21 | xpt, pbuX | Xanthine phosphoribosyltransferase, Xanthine permease | 7.8* | 3.7* | 3.6* |
| SXYL_01690 | pyrP | Uracil permease | 16.2 | 6.3 | 4.1 |
| SXYL_02420 | hpt | Hypoxanthine phosphoribosyltransferase | 6.7 | 4.0 | 2.7 |
| SXYL_00017 | purA | Adenylosuccinate synthetase | 13.2 | 8.6 | 5.0 |
| SXYL_00796 | pdp | Pyrimidine-nucleoside phosphorylase | 2.0 | 2.8 | 2.5 |
| SXYL_00821 | upp | Uracil phosphoribosyltransferase | 9.6 | 8.2 | 6.1 |
| SXYL_01689-85 | pyrBC, carAB, pyrF | UMP biosynthesis | 7.8* | 5.0* | 2.9* |
| SXYL_00807 | pyrG | CTP synthase | 5.7 | 5.2 | 4.3 |
| Division | |||||
| SXYL_01703 | ftsA | Cell division protein FtsA | 2.9 | 2.8 | 2.6 |
| SXYL_01801 | zapA | Cell division protein ZapA | 2.7 | 2.2 | 2.2 |
| SXYL_01704 | divIB | Cell division protein DivIB | 4.8 | 3.5 | 2.7 |
| SXYL_00848 | Cell division protein, FtsW/RodA/SpoVE family | 4.1 | 2.6 | 2.0 | |
| SXYL_01212 | mreD | Cell shape-determining protein MreD | 3.2 | 3.0 | 2.4 |
| SXYL_01651 | smc | Chromosome partition protein Smc | 3.5 | 3.6 | 2.2 |
| SXYL_01699 | sepF | Cell division protein sepF | 0.3 | 0.2 | 0.3 |
| SXYL_01710 | mraZ | Protein MraZ | 0.3 | 0.1 | 0.2 |
| SXYL_00116 | sceD2 | Probable transglycosylase SceD 2 | 34.5 | 31.9 | 49.3 |
| Peptidoglycan synthesis | |||||
| SXYL_00822 | mnaA | UDP-N-acetylglucosamine 2-epimerase | 8.5 | 6.6 | 4.5 |
| SXYL_01425 | murG | UDP-N-acetylglucosamine-tr ansferase | 2.4 | 2.9 | 2.3 |
| SXYL_02467 | sle1 | N-acetylmuramoyl-L-alanine amidase Sle1 | 5.7 | 5.2 | 6.1 |
| SXYL_01706-05 | mraY, murD | Phospho-N-acetylmuramoyl-pentapeptide-transferase, UDP-N-acetylmuramoylalanine–D-glutamate ligase | 8.3* | 5.0* | 3.1* |
| SXYL_01914 | murE | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–L-lysine ligase | 6.2 | 5.2 | 3.6 |
| SXYL_02166 | uppP | Undecaprenyl-diphosphatase | 2.5 | 2.3 | 2.2 |
| SXYL_01624 | uppS | Isoprenyl transferase | 6.1 | 3.0 | 2.5 |
| SXYL_01707 | pbp1 | Penicillin-binding protein 1 | 4.3 | 2.7 | 2.0 |
| SXYL_01304 | pbp3 | Penicillin-binding protein 3 | 2.6 | 3.3 | 2.7 |
| Teichoic acid synthesis | |||||
| SXYL_02208 | tagD | Glycerol-3-phosphate cytidylyltransferase | 0.2 | 0.2 | 0.3 |
| SXYL_01022 | Teichoic acid translocation permease protein | 6.4 | 5.9 | 4.6 | |
| SXYL_01987-90 | dltDCBA | Teichoic acid biosynthesis | 7.6* | 4.7* | 2.6* |
| Membrane proteins | |||||
| SXYL_01650 | ftsY | Signal recognition particle receptor FtsY | 4.2 | 4.8 | 3.0 |
| SXYL_00842 | yidC | Membrane protein insertase YidC | 2.7 | 2.0 | 2.1 |
| SXYL_02419 | ftsH | ATP-dependent zinc metalloprotease FtsH | 11.7 | 12.0 | 8.2 |
| Lipid/Phospholipid synthesis | |||||
| SXYL_01167-68 | accDA | Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta and alpha | 3.6* | 3.7* | 3.0* |
| SXYL_01326-27 | accBC | Acetyl-CoA carboxylase, biotin carboxyl carrier protein and carboxylase subunit | 5.6* | 3.2* | 2.4* |
| SXYL_01655-54 | fabDG | Malonyl CoA-acyl carrier protein transacylase, 3-oxoacyl-[acyl-carrier protein] reductase | 8.9* | 8.6* | 5.1* |
| SXYL_01944-43 | fabHF | 3-oxoacyl-[acyl-carrier-protein] synthase 3 and 2 | 2.7* | 2.6* | 2.1 |
| SXYL_01921 | fabI | Enoyl-[acyl-carrier-protein] reductase [NADPH] | 2.5 | 2.3 | |
| SXYL_01656 | plsX | Phosphate acyltransferase | 13.3 | 9.8 | 6.6 |
| SXYL_01657 | fapR | Transcription factor FapR | 8.0 | 5.1 | 3.2 |
| Regulator/Sensor | |||||
| SXYL_01427-28 | arlSR | Signal transduction histidine-protein kinase ArlS, Regulator ArlR | 2.6* | 3.9* | 2.8* |
| SXYL_00951 | vraS | Sensor protein VraS | 3.8 | 4.6 | 2.7 |
| Lysis | |||||
| SXYL_00365-67 | cidABC | Holin-like proteins CidA and CidB, Pyruvate oxidase | 23.4* | 10.4* | 9.1* |
| SXYL_00494 | lrgA | Antiholin-like protein LrgA | 2.6 | ||
| SXYL_01730-82 | Phage proteins | 13,9* | 14.1* | ||
| SXYL_01785 | Phage repressor-like protein | 0.3 | 0.4 | 0.5 | |
| SXYL_01783 | Phage antirepressor protein | 2.8 | 24.6 | 22.2 | |
| CARBOHYDRATE METABOLISM | |||||
| SXYL_00699 | glcU | Glucose uptake protein | 10.2 | 7.8 | 5.4 |
| Pentose phosphate pathway | |||||
| SXYL_00698 | gdh | Glucose 1-dehydrogenase | 3.0 | 3.2 | 2.8 |
| SXYL_00361 | Gluconolactonase | 3.9 | 3.4 | 2.5 | |
| SXYL_00438-40 | gntRKP | Gluconate Repressor, Kinase, Permease | 19.8* | 38.6* | 16.1* |
| SXYL_00568 | rpi | Ribose-5-phosphate isomerase | 2.5 | 3.0 | |
| EMBDEN-meyerhof-parnas pathway | |||||
| SXYL_01308 | glkA | Glucokinase | 2.8 | 2.9 | |
| SXYL_02069-67 | pgk, tpiA, gpmI | Phosphoglycerate kinase, Triosephosphate isomerase, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | 6.5* | 4.5* | 3.1* |
| Pyruvate metabolism | |||||
| SXYL_00250 | L-lactate permease | 2.5 | 2.6 | ||
| SXYL_00170 | lqo | L-lactate-quinone oxidoreductase | 2.5 | ||
| SXYL_00276 | ldhB | L-lactate dehydrogenase | 0.2 | 0.3 | 0.4 |
| SXYL_01023-24 | pflAB | Formate acetyltransferase | 2.2* | 2.3* | 2.1* |
| SXYL_01839-42 | pdhABCD | Pyruvate dehydrogenase complex | 3.7* | 3.4* | 2.9* |
| SXYL_00194 | lplA | Lipoate-protein ligase A | 6.4 | 11.3 | 6.1 |
| SXYL_02616 | Acetate-CoA ligase | 3.5 | 4.3 | 3.6 | |
| TCA CYCLE AND RESPIRATORY CHAIN | |||||
| SXYL_01793-95 | sdhABC | Succinate dehydrogenase | 3.0* | 4.4* | 3.5* |
| SXYL_01636-37 | sucCD | Succinyl-CoA synthase | 2.5* | 2.2* | |
| SXYL_00579 | mqo | Malate:quinone oxidoreductase | 5.0 | 6.3 | 4.7 |
| SXYL_00824-31 | atpBEFHAGDC | F0F1-type ATP synthase | 8.7* | 5.5* | 4.0* |
| SXYL_00823 | atpI | Putative ATP synthase protein I | 7.3 | 3.7 | 2.6 |
| SXYL_00539-42 | narGHIJ | Respiratory nitrate reductase | 4.2* | 4.4* | 3.1* |
| SXYL_00534-36 | nirBD, sirB | Nitrite reductase | 6.3* | 3.6* | 3.3* |
| SXYL_00547 | narT | Nitrate transporter NarT | 2.3 | ||
| SXYL_00530 | Formate/nitrite transporter | 2.9 | 2.2 | 2.4 | |
| Regulators | |||||
| SXYL_00544 | nreB | Oxygen sensor histidine kinase NreB | 2.1 | ||
| SXYL_01366 | srrB | Sensor histidine kinase SrrB | 2.7 | 2.1 | |
| SXYL_00883 | rex | Redox-sensing transcriptional repressor Rex | 0.4 | 0.4 | 0.4 |
| COFACTOR, VITAMIN SYNTHESIS | |||||
| Molybdenum | |||||
| SXYL_00675-77 | modABC | Molybdate ABC transporter | 5.5* | 5.0* | 3.7* |
| SXYL_00682-87 | moeA, mobAB, moaAE | Molybdenum biosynthesis | 3.0* | 3.1* | 2.9* |
| SXYL_00679 | moeB | Molybdopterin biosynthesis protein MoeB | 2.1 | 1.8 | 1.7 |
| Heme | |||||
| SXYL_01196-99 | hemCDBL1 | Heme biosynthesis | 3.3* | 4.0* | 3.2* |
| SXYL_01274 | hemN | Oxygen-independent coproporphyrinogen oxidase III | 6.6 | 5.0 | 3.2 |
| SXYL_00972 | hemL2 | Glutamate-1-semialdehyde 2,1-aminomutase 2 | 3.1 | 3.3 | 2.5 |
| SXYL_01040-41 | hemHG | Heme biosynthesis | 4.2* | 4.9* | 5.6* |
| SXYL_01819 | ctaB | Protoheme IX farnesyltransferase | 2.8 | 2.5 | |
| SXYL_00751-53 | htsABC | heme transport system | 2.9* | 2.8* | 2.4* |
| SXYL_00755 | isdG | Heme-degrading monooxygenase | 0.2 | 0.2 | 0.3 |
| SXYL_00749 | sfaB | Siderophore biosynthesis protein, IucA/IucC family | 0.1 | 0.2 | 0.2 |
| SXYL_00944 | ftnA | Ferritin | 0.2 | 0.3 | 0.4 |
| Menaquinone | |||||
| SXYL_01079-80 | menEC | Menaquinone biosynthesis | 2.3 | 2.8* | 2.1 |
| SXYL_01891-92 | menHD | Menaquinone biosynthesis | 3.2* | 5.2* | 4.2* |
| Riboflavin | |||||
| SXYL_01097-100 | ribDEBAH | Riboflavin biosynthesis protein | 0.2* | 0.2* | 0.3* |
| Folate | |||||
| SXYL_02414-16 | folKBP | Folate biosynthesis | 5.1* | 5.1* | 3.4* |
| SXYL_01203 | folC | Tetrahydrofolate synthase | 3.6 | 2.2 | |
| SXYL_01872 | folD | Bifunctional protein FolD | 7.3 | 4.6 | 3.3 |
| SXYL_01261 | aroE | Shikimate dehydrogenase | 4.4 | 4.3 | 2.7 |
| PEPTIDE AND AMINO ACID METABOLISM | |||||
| Transport | |||||
| SXYL_00300-02 | Oligopeptide ABC transporter | 2.9* | 2.9* | 2.3* | |
| SXYL_01184 | lysP | Lysine-specific permease | 7.1 | 6.9 | 4.5 |
| SXYL_01528 | Sodium:alanine symporter | 5.11 | |||
| SXYL_01919 | Sodium:alanine symporter | 0.45 | 2.78 | ||
| SXYL_02518 | gltT | Proton/sodium-glutamate symporter | 2.3 | ||
| Peptidase | |||||
| SXYL_01247-48 | U32 family peptidases | 5.8* | 4.5* | 3.2* | |
| SXYL_00957 | ampS | Leucyl aminopeptidase | 3.4 | 2.7 | 2.2 |
| SXYL_00948 | map | Methionine aminopeptidase | 3.0 | 3.0 | 2.0 |
| Valine, leucine, isoleucine | |||||
| SXYL_00867-75 | ilvA, leuDCBA, ilvCNBD1 | Valine, leucine, isoleucine biosynthesis | 0.3* | 0.1* | 0.1* |
| SXYL_02469 | ilvD2 | Dihydroxy-acid dehydratase | 0.2 | 0.1 | 0.1 |
| SXYL_01337-40 | lpdA, bkdA1A2 | alpha-keto acid dehydrogenase complex | 4.9* | 3.7* | 2.8* |
| Tryptophan, phenylalanine, tyrosine | |||||
| SXYL_01383-85 | aroABC | Aromatic acid biosynthesis | 0.3* | 0.3* | 0.2* |
| SXYL_02022 | aroD | 3-dehydroquinate dehydratase | 0.2 | 0.3 | 0.3 |
| SXYL_01497 | trpA | Tryptophan synthase alpha chain | 0.4 | 0.4 | 0.5 |
| SXYL_01128 | DAHP synthetase-chorismate mutase | 0.1 | 0.0 | 0.1 | |
| Histidine | |||||
| SXYL_00460-68 | hisZGDCBHAFI | Histidine biosynthesis | 0.1* | 0.1* | 0.1* |
| Cysteine, methionine | |||||
| SXYL_02644 | metE | 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | 0.1 | 0.1 | 0.2 |
| SXYL_00372 | D-cysteine desulfhydrase | 0.3 | 0.4 | 0.4 | |
| Arginine | |||||
| SXYL_00238-41 | rocD1, argCJB | Arginine biosynthesis | 0.3* | 0.2* | 0.3* |
| SXYL_00252 | arcB | Ornithine carbamoyltransferase | 0.3 | 0.3 | 0.4 |
| SXYL_01961-62 | argGH | Arginine biosynthesis | 0.3* | 0.2* | 0.2* |
| SXYL_01355 | proC | Pyrroline-5-carboxylate reductase | 0.3 | 0.3 | 0.3 |
| Aspartate | |||||
| SXYL_01002 | panD | Aspartate 1-decarboxylase | 0.4 | 0.2 | 0.2 |
| SXYL_01558 | Aspartokinase | 0.4 | 0.3 | 0.4 | |
| SXYL_01373 | asnA | L-asparaginase | 2.6 | 2.8 | 2.4 |
| Glutamate, glutamine | |||||
| SXYL_01964 | gluD1 | Glutamate dehydrogenase | 2.3 | ||
| SXYL_02326 | gluD2 | Glutamate dehydrogenase | 3.5 | ||
| SXYL_02459-61 | gltBCD | Glutamate synthase | 2.6* | ||
| SXYL_00105 | Membrane protein, EutH superfamily | 20.3 | 29.3 | 12.9 | |
| SXYL_00106 | Short-chain dehydrogenase | 41.3 | 29.4 | 13.9 | |
| SXYL_00107 | glnA2 | Glutamine synthetase | 41.7 | 29.2 | 15.7 |
| SXYL_00108 | Aldehyde dehydrogenase | 41.3 | 29.5 | 15.4 | |
| RESPONSE TO OSMOTIC STRESS | |||||
| SXYL_01536 | mscL | Large-conductance mechanosensitive channel | 0.3 | 0.2 | 0.3 |
| Synthesis and accumulation of osmoprotectant | |||||
| SXYL_00488-91 | opuCABCD | Glycine betaine/carnitine/choline ABC transporter | 24.8* | 6.1* | 4.3* |
| SXYL_00486 | lcdH | L-carnitine dehydrogenase | 22.8 | 5.0 | 3.3 |
| SXYL_00223-26 | cudTCA, betA | Glycine betaine/carnitine/choline ABC transporter | 7.9* | 3.7* | 3.4* |
| SXYL_01171 | aapA | D-serine/D-alanine/glycine transporter | 2.6 | 2.8 | 2.3 |
| SXYL_00317 | D-serine/D-alanine/glycine transporter | 4.9 | 6.1 | 4.4 | |
| SXYL_02528-29 | sdaAA1AB1 | L-serine dehydratase, alpha subunit and beta subunit | 3.8* | 5.0* | 4.0* |
| SXYL_00820 | glyA | Serine hydroxymethyltransferase | 10.7 | 7.4 | 5.4 |
| SXYL_01084 | metK | S-adenosylmethionine synthetase | 6.8 | 8.3 | 4.8 |
| SXYL_02526 | Sodium:dicarboxylate symporter | 11.2 | 3.7 | ||
| Na+/H+ antiporter | |||||
| SXYL_01970-76 | mnhA1B1C1D1E1F1G1 | Na(+)/H(+) antiporter | 6.1* | 6.3* | 3.4* |
| SXYL_02220-26 | mnhG2F2E2D2C2B2A2 | Na(+)/H(+) antiporter | 4.8* | 4.6* | 2.8* |
| Regulator | |||||
| SXYL_00859 | rsbU | Serine phosphatase RsbU, regulator of sigma subunit | 4.2 | 2.9 | 2.8 |
| SXYL_00860 | rsbV | Anti-sigma-B factor antagonist | 0.3 | 0.2 | 0.2 |
| SXYL_00861 | rsbW | Serine-protein kinase RsbW | 0.2 | 0.2 | 0.2 |
| STRESS RESPONSE AND PIGMENTATION | |||||
| SXYL_01551 | katB | Catalase B | 0.2 | 0.3 | 0.4 |
| SXYL_02533 | katC | Catalase C | 0.2 | 0.2 | 0.2 |
| SXYL_01572 | bsaA | Glutathione peroxidase | 0.2 | 0.4 | 0.4 |
| SXYL_00374 | Thioredoxin | 0.1 | 0.2 | 0.2 | |
| SXYL_00051-54 | crtPQMN | pigment biosynthesis | 7.8* | 6.3* | 3.9* |
| SXYL_00358 | mvaS | 3-hydroxy-3-methylglutaryl CoA synthase | 2.2 | ||
| SXYL_00359-60 | mvaCA | Acetyl-CoA acetyltransferase, Hydroxymethylglutaryl-CoA reductase | 2.4* | 2.2* | |
| SXYL_02258-56 | mvaK1DK2 | Mevalonate kinase, Diphosphomevalonate decarboxylase, Phosphomevalonate kinase | 5.0* | 4.4* | 2.9* |
| SXYL_00599 | fni | Isopentenyl-diphosphate delta-isomerase | 2.2 | 3.2 | 2.5 |
| SXYL_01332 | ispA | Geranyltranstransferase | 4.6 | 3.8 | 2.5 |
| SXYL_00309 | Universal stress protein | 0.1 | 0.2 | 0.2 | |
| SXYL_01162 | Universal stress protein | 0.2 | 0.2 | 0.2 | |
| SXYL_00196 | General stress protein | 0.0 | 0.0 | 0.0 | |
Means of the expression of the clustered genes differentially expressed.
The response of the information storage group consisted of 60 genes that were mostly up-regulated, 19 of which were involved in DNA replication and recombination, such as polymerases (dnaE, SXYL_01799), topoisomerase IV (parCE), helicases (dnaB, ruvB, recG, recQ1, addA), gyrases (gyrAB) and mismatch repair (mfd, mutS1, mutS2) (Table 1). Thirty genes were involved in translation, such as genes encoding ribosomal proteins, amino-acyl tRNA ligases and proteins involved in synthesis of queuosines, which are modified nucleosides present in certain tRNAs (Supplementary Table 2). Furthermore, the gene fusA encoding the elongation factor G and the gene lepA, a ribosomal elongation factor that recognizes ribosomes after a defective translocation reaction and induces a back-translocation, and two genes infA and infC encoding translation initiation factors were up-regulated (Table 1).
Fourteen genes involved in nucleotide transport and metabolism were up-regulated (Table 1). This metabolism derived from the salvage of preformed extracellular nucleobases as highlighted by pbuX encoding a xanthine permease and pyrP a uracil permease. In meat, ATP is rapidly split into ADP and subsequently into AMP and IMP, which is degraded to inosine and hypoxanthine and further oxidized to xanthine (Battle et al., 2000, 2001). Other nucleotide metabolites are present in pork meat such as uracil (Battle et al., 2000). Together with the gene pbuX, three other genes (xpt, hpt, purA) encoding functions involved in the conversion of purine bases into nucleotides (IMP, XMP, GMP) were overexpressed. Adenine and guanine compounds are interconvertible through the intermediate formation of IMP. Similarly, in addition to pyrP, eight genes (pdp, upp, pyrBC, carAB, pyrF, pyrG) encoding functions involved in the production of UMP and CTP were identified. These genes encode enzymes that belong to either purine or pyrimidine salvage and interconversion pathways as well described for Bacillus subtilis (Switzer et al., 2002). The ability of coagulase-negative staphylococci to metabolize adenosine and inosine in a meat simulation medium has been demonstrated and these compounds may serve as potential alternative energy sources (Janssens et al., 2014).
Eight genes involved in cell division were overexpressed (Table 1). Division requires the participation of numerous proteins that can be functionally discriminated in Z-ring organizing proteins, which are involved in peptidoglycan construction and regulatory proteins (den Blaauwen, 2013). The first step is the polymerization of the FtsZ-ring with a number of Z-ring associated proteins such as FtsA and ZapA encoded by ftsA and zapA. The genes divIB, SXYL-00848 encoding proteins associated with septum synthesis and mreD involved in cell shape were up-regulated. In S. xylosus the genes ftsA and divIB belonged to a cluster of genes governing cell division and peptidoglycan synthesis as already established for several bacteria including S. aureus (Margolin, 2000). The gene smc, which plays a role in chromosome partitioning was up-regulated and two genes (sepF, mraZ) encoding proteins involved in septation and separation of one cell into two daughter cells were down-regulated. Finally, the gene sceD2 encoding a transglycosylase potentially able to cleave peptidoglycan and affect separation of bacterial cells was highly up-regulated.
Concomitantly, 8 genes (mnaA, murG, sle1, mraY, murD, murE, uppP, uppS) involved in peptidoglycan synthesis and two genes (pbp1, pbp3) encoding penicillin-binding proteins involved in its translocation across the membrane were up-regulated (Table 1). The arlSR genes, which encode the two-component regulatory system ArlSR, were up-regulated. An arlS mutant in S. aureus exhibited altered activity of peptidoglycan hydrolases, which participate in important processes occurring during cell growth and division, such as cell wall synthesis, peptidoglycan turnover and recycling (Fournier and Hooper, 2000). Also, vraS encoding the sensor protein of the two-component system VraSR was overexpressed in S. xylosus. In S. aureus, VraSR positively modulates the regulation of the cell wall biosynthesis pathway (Kuroda et al., 2003). The tagD gene encoding the synthesis of teichoic acids was down-regulated in S. xylosus, while the gene SXYL_01022 and the dlt operon encoding, respectively, a permease protein involved in their translocation across the membrane and proteins involved in their alanylation were up-regulated.
Sixteen genes encoding membranous proteins were modulated, with 12 of them up-regulated (Table 1, Supplementary Table 2). Among these, we identified ftsY, which encodes FtsY involved in targeting and insertion of nascent membrane proteins into the cytoplasmic membrane, and yidC, which encodes YidC, which functions as a membrane protein insertase independent of the Sec protein–conducting channel. YidC can also assist in the lateral integration and folding of membrane proteins that insert into the membrane via the Sec pathway (Samuelson et al., 2000). The ftsH gene was involved in the quality control of integral membrane proteins.
In parallel, 13 genes involved in membrane lipid synthesis were up-regulated (Table 1). The mechanism of fatty acid synthesis is conserved and proceeds in two stages: initiation and elongation (Schujman and de Mendoza, 2008). The accDA genes encode the two subunits of carboxyltransferase, while the accBC genes encode the biotin acetyl CoA carboxylase, all of which are involved in the initiation step. In B. subtilis, the transcription of the operon accBC is under growth rate control, the rate of transcription decreasing with decreased growth (Marini et al., 2001). The genes fabDG, fabHF, fabI are involved in the elongation step, while plsx is involved in phospholipid synthesis. PlsX in B. subtilis plays an important role in the coordination of production of fatty acids and phospholipids (Schujman et al., 2003). FapR is a global transcriptional repressor that controls the expression of the fap regulon including all the fab genes and the plsx one (Schujman et al., 2003). The binding of FapR to its DNA targets is specifically inhibited by malonyl-CoA, a cellular pool that provides a mechanism for sensing the status of fatty acid synthesis. In our conditions, up-regulation of acc genes could lead to malonyl-CoA that can control the activity of FapR encoded by fabR up-regulated.
Fifty-one genes involved in cell lysis were modulated (Table 1, Supplementary Table 2). The cidABC cluster was overexpressed. In S. aureus, Cid and Lrg proteins are involved in the control of cell lysis (Ranjit et al., 2011). The S. aureus cid operon expresses two overlapping transcripts: a full-length cidABC transcript that is expressed at low levels during exponential growth and a cidBC transcript also expressed during exponential growth but at high levels (Rice et al., 2004). In S. xylosus, the cidA gene was highly overexpressed by comparison with the cidB and cidC genes (Supplementary Table 2). In S. aureus, cidA encodes a holin-like protein with a positive effect on murein hydrolase activity and lrgA encodes an antiholin-like protein with an inhibitory effect on these enzymes (Ranjit et al., 2011). In S. aureus, the cidA mutant displayed decreased lysis during biofilm formation while the lrgAB mutant was shown to increase lysis (Mann et al., 2009). In our conditions, the very high expression of cidA compared with expression of lrgA led us to suppose that cell lysis occurred during the survival of S. xylosus in the meat model.
Furthermore, a cluster of 46 genes (SXYL_01730-88) encoding phage proteins was modulated with 44 highly up-regulated at 48 and 72 h of incubation (Table 1, Supplementary Table 2). The gene SXYL_01785 encoding a phage repressor-like protein was down-regulated, while the gene SXYL_01783 coding a phage antirepressor protein was highly up-regulated. In Salmonella phage P22, the antirepressor overcomes the repressor protein involved in the maintenance of lysogeny (Levine et al., 1975). Consequently, in our meat model, particularly after 48 and 72 h of incubation, 44 genes involved in phage multiplication reflecting a lytic phase were highly up-regulated. Moreover, we have shown that a lytic phage can be induced by mitomycin C in S. xylosus C2a (data not shown).
Taken together these results (many up-regulated genes involved in DNA machinery, cellular process, cell lysis and the S. xylosus population remaining stable) suggested that a balance between cell division and cell lysis could be observed in our meat model up to 72 h.
Glucose and lactate catabolized simultaneously
The concentrations of glucose added to the meat model and of lactate originating from the meat were close at T0 and decreased similarly during incubation to undetectable levels at 72 h (Table 2). Concomitantly, an increase of acetate was noted in the meat model throughout incubation associated with a small pH decrease of 0.27 after 48 h of incubation. S. xylosus produced mainly acetate as end-product and simultaneously metabolized glucose and lactate in the meat model. It has been shown that S. aureus is able to grow using glucose and lactate simultaneously to form acetate under aerobiosis (Ferreira et al., 2013).
Table 2.
Glucose and lactate consumption and acetate production in a meat model over time.
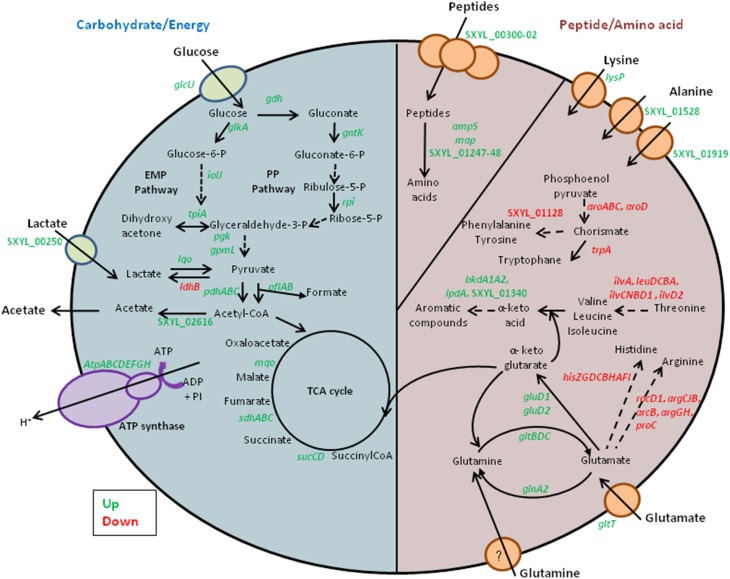
Our transcriptomic data revealed that several genes involved in carbohydrate metabolism were overexpressed (Table 1, Figure 1). Glucose could be transported by the glucose uptake protein (GlcU). Then it could be metabolized via the glucose 1-dehydrogenase (Gdh). In S. xylosus, the genes glcU and gdh are co-localized and co-expressed, suggesting that GlcU recruited glucose for glucose dehydrogenase (Fiegler et al., 1999). GlcU serves in addition to PTS in taking up glucose during S. xylosus growth in laboratory medium (Fiegler et al., 1999). The Gdh and gluconolactonase activities resulted in formation of gluconate and consequently there was overexpression of the cluster gntRKP, encoding the repressor protein, gluconate kinase and gluconate permease, respectively, and involved in gluconate utilization via the pentose phosphate (PP) pathway (Table 1). For Bacillus subtilis, expression of gnt is derepressed in the presence of gluconate (Fujita and Fujita, 1987; Yoshida et al., 1995). In our conditions, gluconate 6-P could be further metabolized via the PP pathway and entered the Embden-Meyerhof-Parnas (EMP) pathway via D-glyceraldehyde 3-P (Table 1, Figure 1). In parallel, the gene glkA encoding glucokinase could fuel the EMP pathway with four genes up-regulated (Table 1). Deviation of part of the catabolism of glucose to the PP pathway could be an advantage as it will generate two NADH contributing to the redox status.
Figure 1.
Summary of the carbohydrate, peptide and amino acid metabolism in Staphylococcus xylosus in a meat model.
Finally, lactate used as carbohydrate substrate could be imported via the L-lactate permease encoded by the gene SXYL_00250, which is overexpressed in the meat model, and catabolized to pyruvate by the lactate-quinone oxidoreductase encoded by lqo, which was also overexpressed (Table 1, Figure 1). S. aureus consumes lactate by a lactate-quinone oxidoreductase that is critical to respiratory growth on L-lactate (Fuller et al., 2011). Interestingly, ldhB encoding lactate dehydrogenase was down-regulated (Table 1, Figure 1). Pyruvate could be catabolized to acetyl-CoA by both formate acetyltransferase and the pyruvate dehydrogenase complex (PDH) (Table 1). Moreover the lplA gene encoding a lipoate-protein ligase A responsible for lipoylation in the presence of exogenous lipoic acid, a factor required for PDH, was also up-regulated (Table 1). Acetyl CoA could therefore be catabolized to acetate by acetate-CoA ligase encoded by SXYL_02616 and excreted in the meat model as early as the first 24 h of incubation (Tables 1, 2).
In our conditions, 100% of the two carbon sources (glucose and lactate) were consumed toward EMP and PP (Table 2, Figure 1). Their catabolism generated acetyl-CoA with 55% (up to 170 mmoles) arising from lactate and 45% (up to 140 mmoles) from glucose. Acetyl-CoA can be then catabolized in acetate or feed the TCA cycle. Only small amounts of acetate were formed (Table 2). So we can hypothesize that the acetyl-CoA to acetate conversion rate did not exceed 14%.
TCA cycle and respiratory chain
Acetyl-CoA is catabolized through the tricarboxylic acid (TCA) cycle, which is a central pathway for the catabolism of carbohydrate, fatty and amino acids, which enter the cycle at several points. Some TCA cycle genes were up-regulated in the meat model, such as the succinate dehydrogenase (sdhABC), succinyl-CoA synthase (sucCD), and malate:quinone oxidoreductase (mqo) genes (Table 1). For S. aureus in biofilm, it was found that the up-regulation of succinate dehydrogenase genes is an advantage under oxygen-limited conditions (Gaupp et al., 2010). The succinate dehydrogenase is involved in the electron transfer of the respiratory chain driving ATP synthesis. We observed that 8 genes encoding the F0F1-type ATP synthase (atpBEFHAGDC) and the gene atpI, upstream of the atp genes and encoding a putative ATP synthase protein I, were highly overexpressed and could furnish energy for the survival of S. xylosus in meat (Table 1, Figure 1).
The overexpression of several genes involved in the EMP and PP pathways, of the clusters nar and nir related to nitrate respiration, although nitrate was not present, of the nitrate and formate/nitrite transporter genes and of pfl encoding formate acetyl transferase, suggested anaerobic conditions (Table 1), as already mentioned for S. aureus grown in vitro in the absence of oxygen (Fuchs et al., 2007). The two-component regulatory system NreBC, which is considered as an oxygen-sensing system, stimulates the expression of genes of nitrate respiration under anaerobic conditions in S. carnosus and S. aureus (Schlag et al., 2008; Reinhart et al., 2010). The targets are the genes encoding the nitrate and nitrite reductases and the nitrate transporter. In our conditions, the regulatory gene nreB was up-regulated at 24 h of growth, thus explaining the up-regulation of nar, nir, and narT (Table 1). Moreover, ssrB encoding a sensor that autophosphorylates in the absence of O2 was up-regulated. This two-component system SrrAB is induced under oxygen-limiting conditions in staphylococci (Green et al., 2014). In S. aureus, a redox-sensing transcriptional repressor Rex is involved in the regulation of anaerobic gene expression (Pagels et al., 2010). The binding activity of Rex is enhanced by NAD+ while NADH decreases it. Rex regulates the expression of pathways that lead to anaerobic NAD+ regeneration, nitrate respiration and ATP synthesis. In our study, the transcription of rex was down-regulated and could explain the overexpression of genes involved in EMP and PP pathways, nitrate respiration and ATP synthesis.
Cofactor, vitamin synthesis
In the meat model, S. xylosus overexpressed the cluster modABC encoding the molybdate ABC-type transporter and six genes, including moeB, of the cluster involved in the synthesis of the molybdenum cofactor (Table 1). In S. carnosus, anaerobic growth conditions enhanced transcription of moeB (Neubauer et al., 1998). Moreover, S. carnosus mutants defective either in modABC or in moeB were defective in nitrate reductase activity, the nitrate reductase being a molybdoenzyme (Neubauer et al., 1999). As we mentioned above, S. xylosus in the meat model also up-regulated the cluster nar encoding the nitrate reductase.
The cluster hemCDBL1 and five other genes (hemN, hemL2, hemHG, ctaB) encoding proteins involved in the synthesis of heme were up-regulated (Table 1). Heme is vital to many biological systems and a multitude of redox enzymes engage heme as a catalyst of electron transfer. S. aureus can employ heme as a cofactor required for respiration, both by aerobic electron transport and by the anaerobic nitrate reductase complex (von Eiff et al., 2006). Moreover, the cluster htsABC encoding a heme transport system was up-regulated in S. xylosus (Table 1). In S. aureus, inactivation of HtsABC reduced its ability to import heme iron (Skaar et al., 2004). Heme can be an iron source, but in the meat model S. xylosus down-regulated the gene isdG encoding a heme-degrading monooxygenase that promotes catalytic degradation of heme with subsequent release of iron. Furthermore, there was down-regulation of sfaB belonging to the cluster sfa involved in synthesis of the siderophore staphyloferrin A and of ftnA involved in iron storage (Table 1). Thus, in the meat model, i.e., in iron-replete conditions, the synthesis of heme is more probably intended for proteins involved in respiration, such as succinate dehydrogenase encoded by sdh (Table 1). The genes menEC and menHD involved in synthesis of menaquinone from chorismate were overexpressed (Table 1). Menaquinone is an important cofactor that is exploited in electron transport pathways. Menaquinone biosynthesis in facultative anaerobes is increased by anaerobiosis and in S. aureus menaquinone deficiency is accompanied by impaired nitrate respiration (Bentley and Meganathan, 1982). During respiration, menaquinone donate electrons to the heme molecules located within cytochromes (Wakeman et al., 2012).
The cluster ribDEBAH involved in riboflavin synthesis was down-regulated in our conditions (Table 1). The active forms of riboflavin in the cell are flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), but riboflavin can be found as free compounds (Perkins and Pero, 2002). In contrast, the cluster folKBP and 3 other genes involved in the synthesis of folate were overexpressed (Table 1). Cells require folate cofactors as acceptor/donor of one carbon unit in numerous processes such as methionine, purine, and thymine synthesis and in some degradative reactions (Rossi et al., 2011).
Peptide and amino acid metabolism
Proteins are the main components of meat. They are classified as water-soluble (sarcoplasmic, 30%), soluble in high salt concentration (myofibrillar, 55%) and insoluble (stromal proteins, proteins from connective tissue) (Lafarga and Hayes, 2014). The sarcoplasmic and myofibrillar proteins are hydrolyzed during fermentation and ripening of fermented sausages (Hughes et al., 2002). The initial degradation is essentially due to the activity of endogenous proteinases that release peptides that can be further hydrolyzed by bacterial enzymes (Hughes et al., 2002). These peptides can be transported into S. xylosus by the oligopeptide transport system encoded by the three genes, SXYL_00300-302 (Table 1, Figure 1). Peptides played a key role in bacterial nutrition; their transport is generally regulated at the transcriptional level by intracellular pools of amino acids and by environmental changes such as anaerobic conditions (Hiron et al., 2007). Two genes (SXYL_01247-48) encoding peptidases from U32 family and the genes ampS and map encoding leucyl- and methionine- aminopeptidases, respectively, were overexpressed. These peptidases will furnish amino acids for S. xylosus (Table 1). The starter cultures (Pediococcus pentosaceus plus S. xylosus or Lactobacillus sakei plus S. carnosus) influenced the proteolytic activity and the release of amino acids in sausages (Hughes et al., 2002; Aro Aro et al., 2010). Moreover, L. sakei overexpressed two dipeptidases in the presence of sarcoplasmic proteins (Fadda et al., 2010).
The meat model contained variable levels of free amino acids with a broad pattern of 19 amino acids (Table 3). The total initial concentration (157 mg/100 g) increased up to 24 h and could be mainly attributed to endogenous meat peptidases. After 24 h there was a decrease of the total free amino acid content. Notably, the level of lysine decreased after 24 h and lysP encoding a lysine-specific permease was overexpressed in S. xylosus (Tables 1, 3). Likewise, the concentration of alanine decreased sharply in the meat model after 48 h of incubation and two genes encoding sodium:alanine symporters were modulated during the incubation (Tables 1, 3). In parallel, in the meat model, S. xylosus down-regulated 26 genes involved in the synthesis of branched-chain amino acids (nine genes belonging to the cluster ilv-leu and the gene ilvD2), aromatic amino acids (aroABC, aroD, trpA, SXYL_01128), histidine (hisZGDCBHAFI) and cysteine-methionine (metE, SXYL_00372) (Table 1, Figure 1). In S. aureus, CodY contributes to the regulation of more than 200 genes including most of the ones listed above (Majerczyk et al., 2010). CodY repression occurs in Bacillus cells growing in a medium containing glucose and amino acids (Shivers and Sonenshein, 2004). Furthermore, S. xylosus down-regulated 8 genes involved in synthesis of arginine (rocD1-argCJB, arcB, argGH, proC) (Table 1). In S. aureus, arginine biosynthesis is regulated through the global regulator CcpA and it was shown that a mutant ccpA facilitates the synthesis of arginine via the urea cycle (Nuxoll et al., 2012).
Table 3.
Concentration of free amino acids in a meat model over time.
| Amino | 0 h | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| Acid | mg/100 g | mg/100 g | mg/100 g | mg/100 g |
| VAL | 7.4 | 8.8 | 8.2 | 5.0 |
| LEU | 8.1 | 10.5 | 9.7 | 5.0 |
| ILE | 3.8 | 3.3 | 2.9 | 1.0 |
| THR | 8.0 | 7.9 | 5.7 | 1.4 |
| TYR | 3.8 | 6.1 | 6.2 | 7.3 |
| PHE | 5.0 | 7.6 | 7.9 | 12.4 |
| HIS | 0.0 | 0.0 | 6.9 | 7.6 |
| ASP | 3.6 | 3.7 | 3.1 | 2.9 |
| ASN | 2.9 | 4.3 | 4.3 | 2.1 |
| GLU | 16.0 | 15.7 | 14.8 | 17.5 |
| GLN | 20.5 | 16.9 | 12.0 | 2.1 |
| SER | 5.4 | 5.4 | 3.5 | 0.8 |
| GLY | 12.8 | 8.7 | 3.6 | 0.0 |
| ALA | 27.6 | 34.5 | 27.3 | 4.0 |
| ARG | 6.7 | 11.3 | 11.4 | 6.1 |
| CYS | 1.4 | 1.1 | 0.8 | 0.6 |
| MET | 3.3 | 4.9 | 5.3 | 6.4 |
| LYS | 16.8 | 19.1 | 11.8 | 7.6 |
| PRO | 3.9 | 5.3 | 6.8 | 5.2 |
| Total | 157.0 | 175.1 | 152.2 | 95.0 |
The genes encoding the synthesis of branched-chain amino acids were down-regulated, whereas four genes lpdA, bkdA1A2, SXYL_01340 organized in a cluster and involved in the catabolism of branched-chain amino acids were up-regulated (Table 1, Figure 1). The two genes (bkdA1A2) encode the subunits of branched-chain alpha-keto acid dehydrogenase E1 involved in the production of 3-methyl butanoyl-CoA, the precursor of 3-methyl butanoic acid (Table 1). The concentrations of leucine, isoleucine and valine decreased in the meat model after 48 h of incubation (Table 3). The pathway leading to the synthesis of 3-methyl butanoic acid, 3-methyl butanal and 3-methyl butanol from the catabolism of leucine has been characterized in S. xylosus (Beck et al., 2004). These aroma compounds and more generally methyl aldehydes, methyl acids and methyl alcohols contribute to the flavor of fermented sausages (Berdagué et al., 1993; Stahnke, 1995; Søndergaard and Stahnke, 2002).
Three genes involved in the aspartate metabolism were modulated in S. xylosus grown in the meat model, with two genes, panD and SXYL_01558, down-regulated, while asnA encoding L-asparaginase was up-regulated (Table 1). The gene asnA coding L-asparaginase in L. sakei was induced during the growth of this bacterium in a meat model and the corresponding mutant ΔasnA2 showed reduced growth in this model, suggesting that asparagine could be a source of nitrogen (Hüfner et al., 2007).
Glutamate and glutamine were found in high concentrations in the meat model (Table 3). In contrast to the concentration of glutamate, which remained stable throughout incubation, the concentration of glutamine decreased sharply in the meat model (Table 3). Glutamate could be imported by the glutamate symporter protein encoded by gltT and catabolized by the glutamate dehydrogenases encoded by gluD1 and gluD2 (Table 1, Figure 1). Glutamate dehydrogenase activity provides α-ketoglutarate, which is required for amino acid transamination, which initiates the conversion of amino acids to aromatic compounds. Glutamate could be synthesized by the glutamate synthase from α-ketoglutarate, an intermediate of TCA cycle, and glutamine. This enzyme is encoded by the gltBCD cluster, which was overexpressed at 24 h of incubation (Table 1, Figure 1). Glutamate serves as the major amino group donor for all nitrogen-containing compounds, as a link between nitrogen and carbon metabolism. The glutamate synthesizing and degrading reactions must be tightly controlled to maintain its homeostasis (Gunka and Commichau, 2012). A cluster of four genes (SXYL_00105-108) was highly overexpressed at the three times of incubation (Table 1). This cluster is uncharacterized, but one of these genes potentially encodes a glutamine synthetase (glnA2). In S. xylosus C2a, another gene encodes a glutamine synthetase, glnA1 located in a cluster with glnR. In B. subtilis, the glnRA operon is expressed in the absence of glutamine (Schreier et al., 1989). In the meat model, glnR and glnA1 were not differentially expressed (Supplementary Table 2). Similarly, two genes encoding glutamine synthetases were identified in Halobacillus halophilus. In this bacterium, the expression of glnA2, but not glnA1, was increased in the presence of NaCl (Saum et al., 2006). The up-regulation of glnA2 and the other genes of the cluster in S. xylosus could be linked to the osmotic stress generated by the presence of salt in the meat model.
Response to osmotic stress generated by added NaCl
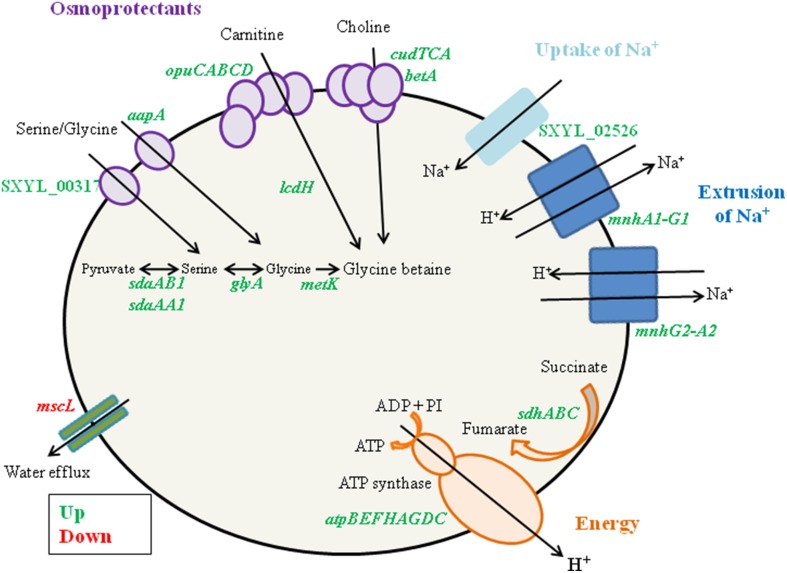
S. xylosus has to adapt to the high concentration of sodium chloride (0.47 M) in the meat model compared with 0.017 M in the medium used for the preparation of the inoculum. A primary response of S. xylosus to the presence of NaCl was the down-regulation of mscL, encoding a large conductance mechanosensitive channel, to prevent water efflux and maintain the physical integrity of the bacterial cells (Table 1, Figure 2).
Figure 2.
Summary of the Staphylococcus xylosus response to osmotic stress generated by NaCl in a meat model.
In addition, genes encoding enzymes involved in three different pathways for the synthesis of glycine betaine, a powerful osmoprotectant, were up-regulated (Table 1, Figure 2). Among them, the cluster opuC (opuCABCD) and lcdH encoding L-carnitine dehydrogenase were identified. The cluster opuC encodes a glycine betaine/carnitine/choline ABC transporter identified as the only uptake route for L-carnitine in B. subtilis and Listeria monocytogenes (Kappes and Bremer, 1998; Angelidis and Smith, 2003). L-carnitine is found in raw meats with levels varying from 6.5 to 87.7 mg/100 g (Demarquoy et al., 2004). In the meat model, L-carnitine could be imported via the OpuC and catabolized to glycine betaine by L-carnitine dehydrogenase. The cluster of four genes (cudTCA, betA) encoding a choline transporter (cudT) and enzymes for dehydrogenation (cudA, betA) to form glycine betaine from choline were also up-regulated. These genes were transcriptionally regulated when S. xylosus C2a was cultured in vitro at high NaCl concentration (Rosenstein et al., 1999). Moreover, the genes aapA and SXYL_00317, which encode D-serine D-alanine glycine transporters, three genes (sdaAA1, sdaAB1, glyA) encoding enzymes involved in the synthesis of glycine, and metK, which encodes S-adenosyl-methionine, which serves as methyl donor for synthesis of glycine betaine, were up-regulated (Table 1, Figure 2). Concomitantly, we observed a sharp decrease of the concentrations of glycine and serine during the incubation in the meat model (Table 3).
The presence of NaCl induced the gene SXYL_02526 encoding a symporter of Na+ and the two clusters mnh (mnhA1-G1, mnhG2-A2) encoding Na+/H+ antiporter systems (Table 1, Figure 2). The modulation of mnh2 expression was previously noted in S. xylosus grown in a meat model supplemented with nitroso compounds, the presence of nitrate and nitrite leading to the down-regulation of mnh2 (Vermassen et al., 2014). In S. aureus, an active antiporter Mnh resulted in increased transcription of genes encoding respiratory chain components, such as the succinate dehydrogenase locus (sdh) (Swartz et al., 2007). The same observation was made in our meat model (Table 1). Moreover, all the genes involved in the synthesis of the multisubunit F0F1-ATPase were overexpressed by S. xylosus, as already mentioned (Table 1, Figure 2). This ATPase links the production of ATP to the transmembrane proton motive force (PMF) and can either generate ATP at the expense of the PMF or generate a PMF using ATP produced by fermentative substrate level phosphorylation (Cotter and Hill, 2003). The PMF can expel the protons, resulting from the activity of the two S. xylosus Mnh antiporters, from the cytoplasm to maintain pH homeostasis. In Bacillus pseudofirmus, proton pumping by the F0F1-ATPase generated a proton motive force across the membrane that powered Mnh antiporter activity after addition of Na+ (Morino et al., 2014).
Sigma factors play a role in the response of bacteria to environmental stress conditions. The expression of rsbU was up-regulated while rsbV and rsbW of the sigma B cluster were down-regulated (Table 1). In S. epidermidis, osmolarity increased the synthesis of polysaccharide intercellular adhesion (PIA) and biofilm formation. The induction of PIA synthesis in the presence of NaCl depends on a functional rsbU gene (Knobloch et al., 2001). Our results with the up-regulation of only rbsU in S. xylosus in the presence of salt could suggest a regulation independent of sigma B.
Response to other stresses
S. xylosus overexpressed the cluster dlt involved in D-alanylation of lipoteichoic and wall teichoic acids (Table 1). The degree of D-alanylation varies depending on environmental conditions such as modification of pH or salt concentration (Neuhaus and Baddiley, 2003). Esterification in S. aureus was shown to increase with a decrease of pH (Neuhaus and Baddiley, 2003). Inactivation of dltC in Streptococcus mutans resulted in the generation of an acid-sensitive strain that could not grow below pH 6.5 (Cotter and Hill, 2003). In the meat model, S. xylosus has to adapt to acidification as the inoculum was grown at pH 7.0 before its inoculation in the meat model at pH 5.9 with a slight decrease during the incubation (Table 2). Thus, the up-regulation of dlt could be the result of adaptation to the acidic environment of meat. It is noteworthy that in the conditions of the meat model, the high concentration of NaCl (0.47 M) did not repress the expression of dlt, as reported for S. aureus in vitro conditions in the presence of 0.325 M NaCl (Koprivnjak et al., 2006).
Growth in the meat model did not seem to generate oxidative stress, as the genes katC, katB, bsaA and SXYL_00374 encoding, respectively, catalases, glutathione peroxidase and thioredoxin involved in the response to this stress, were down-regulated (Table 1). Whereas in our former study, nitrate and nitrate added to the meat model generated nitrosative stress that induced the up-regulation of genes involved in antioxidant defenses (Vermassen et al., 2014).
S. xylosus overexpressed the crtPQMN genes involved in the carotenoid biosynthesis pathway (Table 1). Moreover, eight genes (mvaS, mvaCA, mvaK1DK2, fni, ispA) encoding proteins of mevalonate pathway involved in the synthesis of farnesyl diphosphate, a precursor of the carotenoid synthesis, were up-regulated (Table 1). The C2a strain produces a yellow pigment after cultivation on agar medium, as do about 50% of S. xylosus strains (our unpublished data). S. aureus produced the intermediary yellow pigment 4,4′ diaponeurosporene, which is then converted to the yellow-orange end-product, staphyloxanthin, after prolonged cultivation (Wieland et al., 1994). In S. aureus, the CrtM and CrtN enzymes are responsible for the synthesis of the yellow pigment from farnesyl diphosphate (Wieland et al., 1994). Then, CrtP, CrtQ, CrtO, and AldH catalyze the oxidation, glycosylation and esterification reactions to convert this pigment into staphyloxanthin (Pelz et al., 2005; Kim and Lee, 2012). Staphyloxanthin protects S. aureus against oxidative stress by scavenging free radicals (Clauditz et al., 2006). But more generally carotenoids play a role in overall fitness (Clauditz et al., 2006; Johler et al., 2010) and could contribute to the growth and survival of S. xylosus in meat.
Finally, three genes encoding proteins that are enhanced when the cell is exposed to stress agents were down-regulated (Table 1). All these data suggest that the conditions of growth in meat did not seem to generate stress other than osmotic and acid for S. xylosus.
Conclusion
The global gene expression of S. xylosus in situ in salted meat has allowed us to unravel its adaptation to this complex matrix in conditions that mimic the fermentation period of sausage manufacture. In these conditions, S. xylosus reached a plateau phase after 24 h and a balance between cell division and cell lysis was highlighted during this phase. S. xylosus adapted its metabolism to the meat nutrients and anaerobic conditions. It simultaneously used glucose and lactate as carbon sources. It used peptides and amino acids furnished by the meat. It has to cope essentially with the osmotic stress generated by addition of NaCl. It counteracted this stress by multiple strategies and particularly by the synthesis of glycine betaine, a powerful osmoprotectant.
Author contributions
AV: Contribution to acquisition, analysis and interpretation of the data, contribution in drafting the article. ED: Contribution to experimental and data analysis, critical revising of the manuscript. AD: Statistical data analysis. PM: Contribution to data analysis. VL: Chemical analysis. SL, RT: equally contribute to this paper, design of the work, analysis and interpretation of data, writing of the manuscript
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AV was the recipient of a PhD Research Grant from the French “Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche (M.E.N.E.S.R.).” The microarray development was financially supported by the ANR project “Genoferment” ANR-05-PNRA-020. We would like to thank Muriel Cocaign-Bousquet for helpful discussions and Jean-Paul Chacornac for technical assistance. The authors are grateful to David Marsh for correcting our English.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00087
References
- Angelidis A. S., Smith G. M. (2003). Role of the glycine betaine and carnitine transporters in adaptation of Listeria monocytogenes to chill stress in defined medium. Appl. Environ. Microbiol. 69, 7492–7498. 10.1128/AEM.69.12.7492-7498.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro Aro J. M., Nyam-Osor P., Tsuji K., Shimada K.-I., Fukushima M., Sekikawa M. (2010). The effect of starter cultures on proteolytic changes and amino acid content in fermented sausages. Food Chem. 119, 279–285. 10.1016/j.foodchem.2009.06.025 [DOI] [Google Scholar]
- Barrière C., Centeno D., Lebert A., Leroy-Setrin S., Berdague J. L., Talon R. (2001). Roles of superoxide dismutase and catalase of Staphylococcus xylosus in the inhibition of linoleic acid oxidation. FEMS Microbiol. Lett. 201, 181–185. 10.1016/S0378-1097(01)00271-3 [DOI] [PubMed] [Google Scholar]
- Battle N., Aristoy M.-C., Toldrà F. (2000). Early postmortem detection of exudative pork meat based on nucleotide content. J. Food Sci. 65, 413–416. 10.1111/j.1365-2621.2000.tb16018.x [DOI] [Google Scholar]
- Battle N., Aristoy M.-C., Toldrà F. (2001). ATP metabolites during aging of exudative and nonexudative pork meats. J. Food Sci. 66, 68–71. 10.1111/j.1365-2621.2001.tb15583.x [DOI] [Google Scholar]
- Beck H. C., Hansen A. M., Lauritsen F. R. (2004). Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. J. Appl. Microbiol. 96, 1185–1193. 10.1111/j.1365-2672.2004.02253.x [DOI] [PubMed] [Google Scholar]
- Bentley R., Meganathan R. (1982). Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 46, 241–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdagué J. L., Monteil P., Montel M. C., Talon R. (1993). Effects of starter cultures on the formation of flavour compounds in dry sausage. Meat Sci. 35, 275–287. 10.1016/0309-1740(93)90033-E [DOI] [PubMed] [Google Scholar]
- Clauditz A., Resch A., Wieland K.-P., Peschel A., Götz F. (2006). Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74, 4950–4953. 10.1128/IAI.00204-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. D., Hill C. (2003). Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. 10.1128/MMBR.67.3.429-453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretenet M., Laroute V., Ulve V., Jeanson S., Nouaille S., Even S., et al. (2011). Dynamic analysis of the Lactococcus lactis transcriptome in cheeses made from milk concentrated by ultrafiltration reveals multiple strategies of adaptation to stresses. Appl. Environ. Microbiol. 77, 247–257. 10.1128/AEM.01174-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarquoy J., Georges B., Rigault C., Royer M.-C., Clairet A., Soty M. L., et al. (2004). Radioisotopic determination of l-carnitine content in foods commonly eaten in Western countries. Food Chem. 86, 137–142. 10.1016/j.foodchem.2003.09.023 [DOI] [Google Scholar]
- den Blaauwen T. (2013). Prokaryotic cell division: flexible and diverse. Curr. Opin. Microbiol. 16, 738–744. 10.1016/j.mib.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Derzelle S., Bolotin A., Mistou M.-Y., Rul F. (2005). Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate-lyase as the major upregulated protein. Appl. Environ. Microbiol. 71, 8597–8605. 10.1128/AEM.71.12.8597-8605.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda S., Anglade P., Baraige F., Zagorec M., Talon R., Vignolo G., et al. (2010). Adaptive response of Lactobacillus sakei 23K during growth in the presence of meat extracts: a proteomic approach. Int. J. Food Microbiol. 142, 36–43. 10.1016/j.ijfoodmicro.2010.05.014 [DOI] [PubMed] [Google Scholar]
- Ferreira M. T., Manso A. S., Gaspar P., Pinho M. G., Neves A. R. (2013). Effect of oxygen on glucose metabolism: utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR Studies. PLoS ONE 8:e58277. 10.1371/journal.pone.0058277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H., Bassias J., Jankovic I., Brückner R. (1999). Identification of a gene in Staphylococcus xylosus encoding a novel glucose uptake protein. J. Bacteriol. 181, 4929–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H., Brückner R. (1997). Identification of the serine acetyltransferase gene of Staphylococcus xylosus. FEMS Microbiol. Lett. 148, 181–187. 10.1111/j.1574-6968.1997.tb10286.x [DOI] [PubMed] [Google Scholar]
- Fournier B., Hooper D. C. (2000). A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182, 3955–3964. 10.1128/JB.182.14.3955-3964.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S., Pané-Farré J., Kohler C., Hecker M., Engelmann S. (2007). Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189, 4275–4289. 10.1128/JB.00081-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita T. (1987). The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. Proc. Natl. Acad. Sci. U.S.A. 84, 4524–4528. 10.1073/pnas.84.13.4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller J. R., Vitko N. P., Perkowski E. F., Scott E., Khatri D., Spontak J. S., et al. (2011). Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell. Infect. Microbiol. 1:19. 10.3389/fcimb.2011.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire V., Piot M., Camier B., Vissers J.-P., Jan G., Léonil J. (2004). Survey of bacterial proteins released in cheese: a proteomic approach. Int. J. Food Microbiol. 94, 185–201. 10.1016/j.ijfoodmicro.2004.01.010 [DOI] [PubMed] [Google Scholar]
- Gaupp R., Schlag S., Liebeke M., Lalk M., Götz F. (2010). Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms. J. Bacteriol. 192, 2385–2394. 10.1128/JB.01472-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Rolfe M. D., Smith L. J. (2014). Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide. Virulence 5, 794–809. 10.4161/viru.27794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunka K., Commichau F. M. (2012). Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol. Microbiol. 85, 213–224. 10.1111/j.1365-2958.2012.08105.x [DOI] [PubMed] [Google Scholar]
- Hiron A., Borezée-Durant E., Piard J.-C., Juillard V. (2007). Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189, 5119–5129. 10.1128/JB.00274-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüfner E., Markieton T., Chaillou S., Crutz-Le Coq A. M., Zagorec M., Hertel C. (2007). Identification of Lactobacillus sakei genes induced during meat fermentation and their role in survival and growth. Appl. Environ. Microbiol. 73, 2522–2531. 10.1128/AEM.02396-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. C., Kerry J. P., Arendt E. K., Kenneally P. M., McSweeney P. L., O'Neill E. E. (2002). Characterization of proteolysis during the ripening of semi-dry fermented sausages. Meat Sci. 62, 205–216. 10.1016/S0309-1740(01)00248-0 [DOI] [PubMed] [Google Scholar]
- Janssens M., Van der Mijnsbrugge A., Sánchez Mainar M., Balzarini T., De Vuyst L., Leroy F. (2014). The use of nucleosides and arginine as alternative energy sources by coagulase-negative staphylococci in view of meat fermentation. Food Microbiol. 39, 53–60. 10.1016/j.fm.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Johler S., Stephan R., Hartmann I., Kuehner K. A., Lehner A. (2010). Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress. Appl. Environ. Microbiol. 76, 1053–1061. 10.1128/AEM.01420-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes R. M., Bremer E. (1998). Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine and gamma-butyrobetaine via the ABC transport system OpuC. Microbiology 144, 83–90. 10.1099/00221287-144-1-83 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lee P. C. (2012). Functional expression and extension of staphylococcal staphyloxanthin biosynthetic pathway in Escherichia coli. J. Biol. Chem. 287, 21575–21583. 10.1074/jbc.M112.343020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch J. K., Bartscht K., Sabottke A., Rohde H., Feucht H. H., Mack D. (2001). Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183, 2624–2633. 10.1128/JB.183.8.2624-2633.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivnjak T., Mlakar V., Swanson L., Fournier B., Peschel A., Weiss J. P. (2006). Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 188, 3622–3630. 10.1128/JB.188.10.3622-3630.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Kuroda H., Oshima T., Takeuchi F., Mori H., Hiramatsu K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49, 807–821. 10.1046/j.1365-2958.2003.03599.x [DOI] [PubMed] [Google Scholar]
- Lafarga T., Hayes M. (2014). Bioactive peptides from meat muscle and by-products: generation, functionality and application as functional ingredients. Meat Sci. 98, 227–239. 10.1016/j.meatsci.2014.05.036 [DOI] [PubMed] [Google Scholar]
- Leroy F., Verluyten J., De Vuyst L. (2006). Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 106, 270–285. 10.1016/j.ijfoodmicro.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Levine M., Truesdell S., Ramakrishnan T., Bronson M. J. (1975). Dual control of lysogeny by bacteriophage P22: an antirepressor locus and its controlling elements. J. Mol. Biol. 91, 421–438. 10.1016/0022-2836(75)90270-3 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Majerczyk C. D., Dunman P. M., Luong T. T., Lee C. Y., Sadykov M. R., Somerville G. A., et al. (2010). Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877. 10.1128/JB.00220-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E. E., Rice K. C., Boles B. R., Endres J. L., Ranjit D., Chandramohan L., et al. (2009). Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4:e5822. 10.1371/journal.pone.0005822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. (2000). Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24, 531–548. 10.1111/j.1574-6976.2000.tb00554.x [DOI] [PubMed] [Google Scholar]
- Marini P. E., Perez C. A., de Mendoza D. (2001). Growth-rate regulation of the Bacillus subtilis accBC operon encoding subunits of acetyl-CoA carboxylase, the first enzyme of fatty acid synthesis. Arch. Microbiol. 175, 234–237. 10.1007/s002030100256 [DOI] [PubMed] [Google Scholar]
- Morino M., Suzuki T., Ito M., Krulwich T. A. (2014). Purification and functional reconstitution of a seven-subunit Mrp-type Na+/H+ antiporter. J. Bacteriol. 196, 28–35. 10.1128/JB.01029-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer H., Pantel I., Götz F. (1998). Characterization of moeB- part of the molybdenum cofactor biosynthesis gene cluster in Staphylococcus carnosus. FEMS Microbiol. Lett. 164, 55–62. 10.1016/s0378-1097(98)00196-7 [DOI] [PubMed] [Google Scholar]
- Neubauer H., Pantel I., Lindgren P. E., Götz F. (1999). Characterization of the molybdate transport system ModABC of Staphylococcus carnosus. Arch. Microbiol. 172, 109–115. 10.1007/s002030050747 [DOI] [PubMed] [Google Scholar]
- Neuhaus F. C., Baddiley J. (2003). A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 686–723. 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouaille S., Even S., Charlier C., Le Loir Y., Cocaign-Bousquet M., Loubière P. (2009). Transcriptomic response of Lactococcus lactis in mixed culture with Staphylococcus aureus. Appl. Environ. Microbiol. 75, 4473–4482. 10.1128/AEM.02653-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuxoll A. S., Halouska S. M., Sadykov M. R., Hanke M. L., Bayles K. W., Kielian T., et al. (2012). CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog. 8:e1003033. 10.1371/journal.ppat.1003033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen P. T., Stahnke L. H. (2004). The influence of environmenatl parameters on the catabolism of branched-chain amino acids by Staphylococcus xylosus and Staphylococcus carnosus. Food Microbiol. 21, 43–50. 10.1016/S0740-0020(03)00048-0 [DOI] [Google Scholar]
- Pagels M., Fuchs S., Pané-Farré J., Kohler C., Menschner L., Hecker M., et al. (2010). Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76, 1142–1161. 10.1111/j.1365-2958.2010.07105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz A., Wieland K.-P., Putzbach K., Hentschel P., Albert K., Götz F. (2005). Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280, 32493–32498. 10.1074/jbc.M505070200 [DOI] [PubMed] [Google Scholar]
- Perkins J. B., Pero J. (2002). Vitamin biosynthesis, in Bacillus subtilis and Its Closest Relatives from Genes to Cells, eds Sonenshein A. L., Hoch J. A., Losick R. (New York, NY: ASM Press; ), 271–286. [Google Scholar]
- Ranjit D. K., Endres J. L., Bayles K. W. (2011). Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J. Bacteriol. 193, 2468–2476. 10.1128/JB.01545-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravyts F., Vuyst L. D., Leroy F. (2012). Bacterial diversity and functionalities in food fermentations. Eng. Life Sci. 12, 356–367. 10.1002/elsc.201100119 [DOI] [Google Scholar]
- Reinhart F., Huber A., Thiele R., Unden G. (2010). Response of the oxygen sensor NreB to air in vivo: Fe-S-containing NreB and Apo-NreB in aerobically and anaerobically growing Staphylococcus carnosus. J. Bacteriol. 192, 86–93. 10.1128/JB.01248-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice K. C., Patton T., Yang S.-J., Dumoulin A., Bischoff M., Bayles K. W. (2004). Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 186, 3029–3037. 10.1128/JB.186.10.3029-3037.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein R., Futter-Bryniok D., Götz F. (1999). The choline-converting pathway in Staphylococcus xylosus C2A: genetic and physiological characterization. J. Bacteriol. 181, 2273–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein R., Nerz C., Biswas L., Resch A., Raddatz G., Schuster S., et al. (2009). Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75, 811–822. 10.1128/AEM.01982-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M., Amaretti A., Raimondi S. (2011). Folate production by probiotic bacteria. Nutrients 3, 118–134. 10.3390/nu3010118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., et al. (2000). YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641. 10.1038/35020586 [DOI] [PubMed] [Google Scholar]
- Saum S. H., Sydow J. F., Palm P., Pfeiffer F., Oesterhelt D., Müller V. (2006). Biochemical and molecular characterization of the biosynthesis of glutamine and glutamate, two major compatible solutes in the moderately halophilic bacterium Halobacillus halophilus. J. Bacteriol. 188, 6808–6815. 10.1128/JB.00781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag S., Fuchs S., Nerz C., Gaupp R., Engelmann S., Liebeke M. (2008). Characterization of the oxygen responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 190, 7847–7858. 10.1128/JB.00905-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier H. J., Brown S. W., Hirschi K. D, Nomellini J. F, Sonenshein A. L. (1989). Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J. Mol. Biol. 210, 51–63. 10.1016/0022-2836(89)90290-8 [DOI] [PubMed] [Google Scholar]
- Schujman G. E., de Mendoza D. (2008). Regulation of type II fatty acid synthase in Gram-positive bacteria. Curr. Opin. Microbiol. 11, 148–152. 10.1016/j.mib.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Schujman G. E., Paoletti L., Grossman A. D., de Mendoza D. (2003). FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell. 4, 663–672. 10.1016/S1534-5807(03)00123-0 [DOI] [PubMed] [Google Scholar]
- Shivers R. P., Sonenshein A. L. (2004). Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53, 599–611. 10.1111/j.1365-2958.2004.04135.x [DOI] [PubMed] [Google Scholar]
- Skaar E. P., Humayun M., Bae T., DeBord K. L., Schneewind O. (2004). Iron-source preference of Staphylococcus aureus infection. Science 305, 1626–1628. 10.1126/science.1099930 [DOI] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1–25. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Søndergaard A. K., Stahnke L. H. (2002). Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum, a comparative study in model systems. Int. J. Food Microbiol. 75, 99–109. 10.1016/S0168-1605(01)00729-2 [DOI] [PubMed] [Google Scholar]
- Stahnke L. H. (1995). Dried sausages fermented with Staphylococcus xylosus at different temperatures and with different ingredient levels - Part II. Volatile components. Meat Sci. 41, 193–209. 10.1016/0309-1740(94)00069-J [DOI] [PubMed] [Google Scholar]
- Swartz T. H., Ito M., Ohira T., Natsui S., Hicks D. B., Krulwich T. A. (2007). Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J. Bacteriol. 189, 3081–3090. 10.1128/JB.00021-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer R. L., Zalkin H., Saxild H. H. (2002). Purine, pyrimidine, and pyridine nucleotide metabolism, in Bacillus subtilis and Its Closest Relatives from Genes to Cells, eds Sonenshein A. L., Hoch J. A., Losick R. (New York, NY: ASM Press; ), 255–269. [Google Scholar]
- Taïbi A., Dabour N., Lamoureux M., Roy D., LaPointe G. (2011). Comparative transcriptome analysis of Lactococcus lactis subsp. cremoris strains under conditions simulating Cheddar cheese manufacture. Int. J. Food Microbiol. 146, 263–275. 10.1016/j.ijfoodmicro.2011.02.034 [DOI] [PubMed] [Google Scholar]
- Talon R., Leroy S. (2006). Latest developments in meat bacterial starters, in Advanced Technologies for Meat Processing, eds Nollet L. M. L., Toldrà F. (New York, NY: Taylor and Francis; Group; CRC Press; ), 401–418. [Google Scholar]
- Tjener K., Stahnke L. H., Andersen L., Martinussen J. (2004). The pH-unrelated influence of salt, temperature and manganese on aroma formation by Staphylococcus xylosus and Staphylococcus carnosus in a fermented meat model system. Int. J. Food Microbiol. 97, 31–42. 10.1016/j.ijfoodmicro.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Toldrá F. (2007). Biochemistry of meat and fat, in Handbook of Fermented Meat and Poultry, ed Toldrá F. (Oxford: Blackwell Publishing; ), 51–58. [Google Scholar]
- Vermassen A., de la Foye A., Loux V., Talon R., Leroy S. (2014). Transcriptomic analysis of Staphylococcus xylosus in the presence of nitrate and nitrite in meat reveals its response to nitrosative stress. Front. Microbio. 5:691. 10.3389/fmicb.2014.00691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff C., McNamara P., Becker K., Bates D., Lei X.-H., Ziman M., et al. (2006). Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriol. 188, 687–693. 10.1128/JB.188.2.687-693.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman C. A., Hammer N. D., Stauff D. L., Attia A. S., Anzaldi L. L., Dikalov S. I., et al. (2012). Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 86, 1376–1392. 10.1111/mmi.12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland B., Feil C., Gloria-Maercker E., Thumm G., Lechner M., Bravo J.-M., et al. (1994). Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4′-diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 176, 7719–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Ohmori H., Miwa Y., Fujita Y. (1995). Bacillus subtilis gnt repressor mutants that diminish gluconate-binding ability. J. Bacteriol. 177, 4813–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.