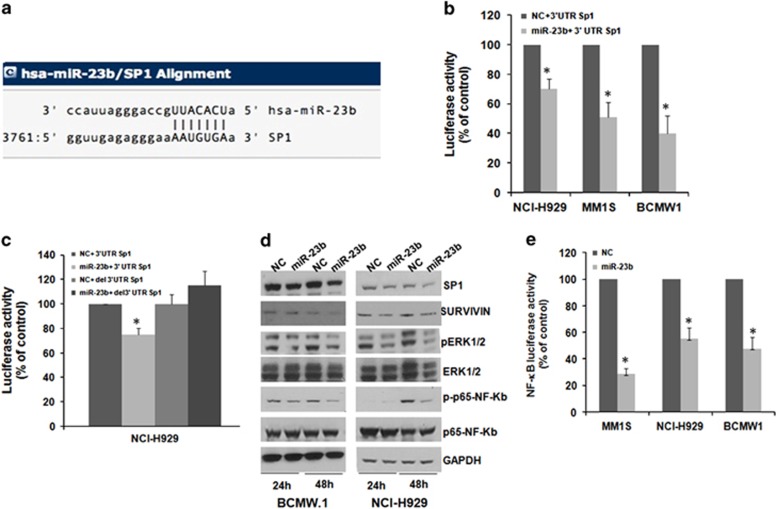
Figure 1.
Sp1 targeting by miR-23b in MM and WM tumor cells. (a) Identification of a miR-23b-target sequence within Sp1 3′UTR by microRNA.org in silico target prediction. (b) Dual-luciferase assay of NCI-H929, MM1s and BCMW1 cells co-transfected with firefly luciferase constructs containing a portion of the 3′UTR of Sp1 (nts 2503–5200) and 200 nm of miR-23b or scrambled oligonucleotides (NC). The firefly luciferase activity was normalized to renilla luciferase activity. The data are shown as relative luciferase activity of miR-23b-transfected cells as compared with the control (NC) of a total of six experiments from three independent transfections. *P<0.01. (c) Dual-luciferase assay of NCI-H929 cells co-transfected with firefly luciferase constructs containing the 3′UTR of Sp1 or a deletion mutant lacking the predicted miR-23b target site (3′UTR del) and 200 nm of miR-23b or scrambled oligonucleotides (NC) as indicated. The firefly luciferase activity was normalized to renilla luciferase activity. *P<0.01. (d) Immunoblot analysis of Sp1, survivin, phosphorylated ERK1/2, phosphorylated p65, p65 in NCI-H929 and BCMW1 cells transfected with 200 nm miR-23b mimics or NC. GAPDH was used as a loading control. (e) Luciferase assay of MM1s, NCI-H929 and BCMW1 cells transfected with 200 nm of miR-23b mimics or NC and a firefly luciferase vector carrying the NF-κB-responsive elements. Renilla luciferase from pCMV-RL was used for normalization. Data are expressed as the ratio of firefly and renilla luciferase and are the mean of three different experiments performed in triplicate. *P<0.01. Abbreviation: GAPDH, glyceraldehyde-3-phosphate dehydrogenase.