ABSTRACT
The upper respiratory tract is colonized by a diverse array of commensal bacteria that harbor potential pathogens, such as Streptococcus pneumoniae. As long as the local microbial ecosystem—also called “microbiome”—is in balance, these potentially pathogenic bacterial residents cause no harm to the host. However, similar to macrobiological ecosystems, when the bacterial community structure gets perturbed, potential pathogens can overtake the niche and cause mild to severe infections. Recent studies using next-generation sequencing show that S. pneumoniae, as well as other potential pathogens, might be kept at bay by certain commensal bacteria, including Corynebacterium and Dolosigranulum spp. Bomar and colleagues are the first to explore a specific biological mechanism contributing to the antagonistic interaction between Corynebacterium accolens and S. pneumoniae in vitro [L. Bomar, S. D. Brugger, B. H. Yost, S. S. Davies, K. P. Lemon, mBio 7(1):e01725-15, 2016, doi:10.1128/mBio.01725-15]. The authors comprehensively show that C. accolens is capable of hydrolyzing host triacylglycerols into free fatty acids, which display antipneumococcal properties, suggesting that these bacteria might contribute to the containment of pneumococcus. This work exemplifies how molecular epidemiological findings can lay the foundation for mechanistic studies to elucidate the host-microbe and microbial interspecies interactions underlying the bacterial community structure. Next, translation of these results to an in vivo setting seems necessary to unveil the magnitude and importance of the observed effect in its natural, polymicrobial setting.
TEXT
The human upper respiratory tract, including the anterior nares, naso- and oropharynx, forms the vestibule to the lower respiratory tract and is colonized by a myriad of bacteria, including both potential pathogens and harmless commensals. Even within an organ system like the respiratory tract, the ecological niches vary according to mucosal architecture, local host immune response, and environmental exposure (i.e., temperature, humidity, and nutrient and oxygen availability), consequently selecting for bacterial species that are capable of withstanding the site-specific conditions. The bacterial community structure of these microhabitats is further defined by bacterial interactions, which can be antagonistic, commensal, or mutual in nature, altogether supporting a healthy equilibrium within the microbiome and between microbiome and host.
For most of research history, studies on the pathogenesis and prevention of acute infectious respiratory diseases were restricted to investigations of pathogenic traits and pathogen colonization of the upper respiratory tract as the first steps in the process leading to respiratory disease (1). However, the advent of next-generation sequencing has made it possible to study pathogens in the context of their complex bacterial communities and, since then, information on the role of both culturable and nonculturable microorganisms in health and disease has been accumulating rapidly. Recent insights suggest an important role of the human microbiome in metabolism (2), resilience to pathogen invasion (3), and the development of the host immune system (4). However, disturbance of the ecological equilibrium by internal or external triggers may lead to loss of homeostasis, overgrowth, and dissemination of potential pathogens, subsequently resulting in local or systemic acute or chronic diseases (4, 5).
Next-generation sequencing of bacterial communities in the upper respiratory tract early in life revealed that the human nasopharyngeal microbiota can adopt a limited number of compositions (6, 7). These bacterial community structures were uniformly observed in geographically distinct Western populations, and thus likely arise from deterministic rather than stochastic (ecological) processes. Interestingly, in all these studies, the presence and abundance of Corynebacterium spp. appears to be negatively associated with pneumococcal carriage and acute respiratory infections, such as acute otitis media (6, 8, 9). These molecular epidemiological findings are highly important in shaping our view of the potential role of the respiratory microbiome in infection and inflammation. However, the direct and indirect underlying biological mechanisms need to be deciphered.
The study of Bomar and colleagues (10) that was recently published in mBio serves as an excellent example demonstrating how the identification of specific bacterial interactions by next-generation sequencing approaches can spark new hypotheses and direct mechanistic studies. The investigators first verify the previously reported negative association between Corynebacterium spp. and Streptococcus pneumoniae—both frequent inhabitants of the infant anterior nares and nasopharynx—in vivo, subsequently studying their interaction in an in vitro setting. They comprehensively show that Corynebacterium accolens inhibits pneumococcal growth by converting human skin-associated triacylglycerols (TAGs) to free fatty acids (FFAs), such as oleic and linoleic acid, which are known to exert antimicrobial activity.
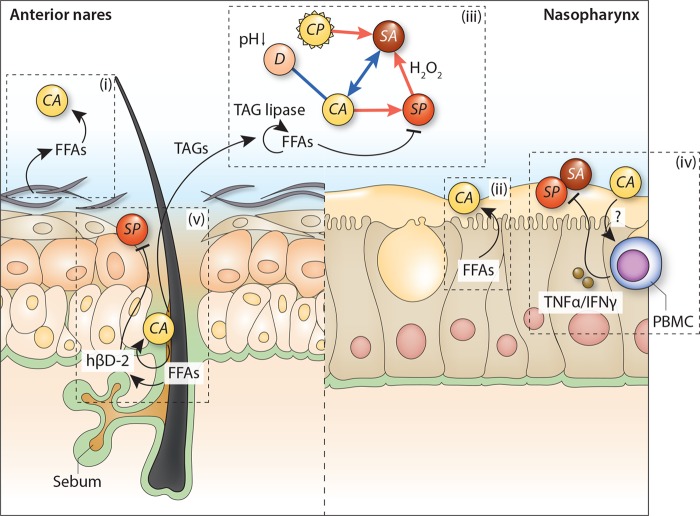
The results of Bomar et al. suggest that the negative association between the presence of Corynebacterium spp. and pneumococcal carriage observed in vivo is mainly deployed through a direct antagonistic interaction between these bacterial species. Although this is likely at least partly true, one could argue that the observed interaction is presumably the result of (multiple) additional biological pathways. For example, previous epidemiological data show that Corynebacterium spp. commonly co-occur with Dolosigranulum spp. and that their joint presence is strongly and negatively associated with S. pneumoniae abundance and a reduced risk of acute otitis media (8, 9). Recently, we speculated that the lactic acid-producing ability of Dolosigranulum spp. might acidify the local environment and select for Corynebacterium spp. outgrowth, explaining their common co-occurrence (5). The question of whether one of these species or both species contribute to the exclusion of potentially pathogenic bacteria from the niche remains highly relevant for the design of potential new preventive strategies (Fig. 1).
FIGURE 1 .
The antagonistic relationship between Corynebacterium accolens and Streptococcus pneumoniae in the context of other known ecological interactions. Growth of C. accolens (CA) is stimulated by (host-derived) free fatty acids (FFAs) that are present throughout the nose in sebum, at the skin surface (i), and in nasal fluid (ii). Additionally, triacylglycerol (TAG) lipase activity of C. accolens results in the release of (microbiota-derived) FFAs, inhibiting the growth of S. pneumoniae (SP) (iii). Other studies suggest an indirect effect of C. accolens on pneumococcal growth through the release of tumor necrosis factor alpha (TNF-α) and interferon gamma (IFN-γ) by peripheral blood mononuclear cells (PBMC) (iv) or by inducing the release of human beta-defensin-2 (hβD-2) from human sebocytes (v). In turn, C. accolens, contrary to Corynebacterium pseudodiphtheriticum (CP), is positively associated with Dolosigranulum spp. (D) and Staphylococcus aureus (SA). The presence of C. accolens may thus (partly) explain the well-established negative association between S. aureus and S. pneumoniae (iii). Host-microbe interactions are depicted using black lines (arrowhead, positive effect; blunt head, negative effect). Microbe-microbe interactions are represented by blue (positive association) and red (negative association) lines. Question marks adjacent to arrows indicate uncertain effects.
Furthermore, in contrast to the potential inhibitory effects on S. pneumoniae, C. accolens is positively associated with the presence and outgrowth of Staphylococcus aureus—like S. pneumoniae a Gram-positive potential pathogen—and vice versa (11). Intriguingly, despite observing this interaction in the absence of FFAs, this alludes to the previously observed negative correlation between S. pneumoniae and S. aureus (12), for which the exact molecular basis in vivo remains unclear (13–15) but which might in fact be partially driven by C. accolens (Fig. 1). Of note here is the finding that the closely related Corynebacterium pseudodiphtheriticum elicits an opposite effect on S. aureus growth, underlining the need for more detailed information on species- and strain-level interactions to explain the more generalized observations from molecular epidemiological studies (11).
Additionally, the observed antagonistic relationship between Corynebacterium spp. and pneumococcus might be shaped by the host immune system as well. Unfortunately, current literature on the potential immunomodulatory capacity of Corynebacterium spp. is sparse. Indirect evidence obtained from patients suffering from IL-17 immune defects suggests that a decreased abundance of corynebacteria within the skin microbiome is associated with a reduced capacity to produce proinflammatory cytokines (tumor necrosis factor alpha [TNF-α] and interferon gamma [IFN-γ]), with a pivotal effect in the defense against several known respiratory pathogens (16). Moreover, as stated by Bomar et al. (10), the Corynebacterium-induced liberation of specific FFAs might lead human sebocytes to release human beta-defensin 2, an antimicrobial peptide that has been demonstrated to exert activity against S. pneumoniae (Fig. 1) (17).
Interestingly, the negative correlation between Corynebacterium spp. and S. pneumoniae was observed in the anterior nares as well as the nasopharynx (7–9), the latter of which is traditionally seen as the niche from which pathobionts disperse to the lungs (1). However, these niches show some major differences with regard to epithelial cell type, humidity, and acidity. Since, in contrast to the nasopharynx, the vestibulum of the nose is covered with skinlike keratinized squamous epithelium containing sebaceous glands, one would expect a preference of the lipophilic Corynebacterium spp. for the sebum-rich anterior nares (18). It is, however, highly likely that the pseudostratified ciliated columnar epithelium lining the nasopharynx also secretes lipid-rich fluids, including oleic and linoleic acids, as was observed by Do and colleagues for primary human bronchoepithelial cells (19). The presumably high concentration of host-derived lipids and FFAs throughout the nose may explain the lack of a biogeographical preference of Corynebacterium spp. for colonizing the nasal cavity, though it introduces the conundrum of the exact additive effect of Corynebacterium-induced liberation of FFAs to the suppression of pneumococcal growth in vivo (Fig. 1).
In conclusion, we strongly agree with the authors that molecular epidemiological approaches for studying the role of the respiratory microbiome in health and disease should be extended with studies identifying the potential molecular mechanisms that underlie the observed bacterial community configurations, and we believe their work is exemplary in that regard. The negative correlation between Corynebacterium spp. and S. pneumoniae, which was previously observed in epidemiological studies, was convincingly mirrored in vitro by Bomar et al., demonstrating that corynebacteria hydrolyze TAGs, leading to the liberation of FFAs and subsequent inhibition of pneumococcal growth. These results provoke speculations on the possible beneficial role of commensal corynebacteria to human health, yet the importance of this mechanism in vivo remains an open question and requires the development of biologically more complex in vitro or in vivo models to properly assess the interaction in its authentic ecological context.
The views expressed in this Commentary do not necessarily reflect the views of this journal or of ASM.
Footnotes
Citation de Steenhuijsen Piters WAA, Bogaert D. 2016. Unraveling the molecular mechanisms underlying the nasopharyngeal bacterial community structure. mBio 7(1):e00009-16. doi:10.1128/mBio.00009-16.
REFERENCES
- 1.Bogaert D, De Groot R, Hermans P. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 2.Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 3.Kamada N, Chen GY, Inohara N, Núñez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Steenhuijsen Piters WAA, Sanders EAM, Bogaert D. 2015. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc B 370:20140294. doi: 10.1098/rstb.2014.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, Bochkov YA, Grindle K, Johnston SL, Gern JE, Sly PD, Holt PG, Holt KE, Inouye M. 2015. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. 2014. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 8.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. 2012. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. 2011. Microbial communities of the upper respiratory tract and otitis media in children. mBio 2:e00245-10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. 2016. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7(1):e01725-15. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho D-Y, Holmes S, Relman DA. 2013. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogaert D, van Belkum A, Sluijter M, Luijendijk A, De Groot R, Rümke HC, Verbrugh HA, Hermans PW. 2004. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 13.Regev-Yochay G, Malley R, Rubinstein E, Raz M, Dagan R, Lipsitch M. 2008. In vitro bactericidal activity of Streptococcus pneumoniae and bactericidal susceptibility of Staphylococcus aureus strains isolated from cocolonized versus noncocolonized children. J Clin Microbiol 46:747–749. doi: 10.1128/JCM.01781-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margolis E. 2009. Hydrogen peroxide-mediated interference competition by Streptococcus pneumoniae has no significant effect on Staphylococcus aureus nasal colonization of neonatal rats. J Bacteriol 191:571–575. doi: 10.1128/JB.00950-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regev-Yochay G, Trzciński K, Thompson CM, Malley R, Lipsitch M. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smeekens SP, Huttenhower C, Riza A, van de Veerdonk FL, Zeeuwen PLJM, Schalkwijk J, van der Meer JWM, Xavier RJ, Netea MG, Gevers D. 2014. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J Innate Immun 6:253–262. doi: 10.1159/000351912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatsuji T, Kao MC, Zhang L, Zouboulis CC, Gallo RL, Huang CM. 2010. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating beta-defensin-2 expression. J Invest Dermatol 130:985–994. doi: 10.1038/jid.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael-Jubeli R, Bleton J, Baillet-Guffroy A. 2011. High-temperature gas chromatography-mass spectrometry for skin surface lipids profiling. J Lipid Res 52:143–151. doi: 10.1194/jlr.D008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Do TQ, Moshkani S, Castillo P, Anunta S, Pogosyan A, Cheung A, Marbois B, Faull KF, Ernst W, Chiang SM, Fujii G, Clarke CF, Foster K, Porter E. 2008. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J Immunol 181:4177–4187. doi: 10.4049/jimmunol.181.6.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]