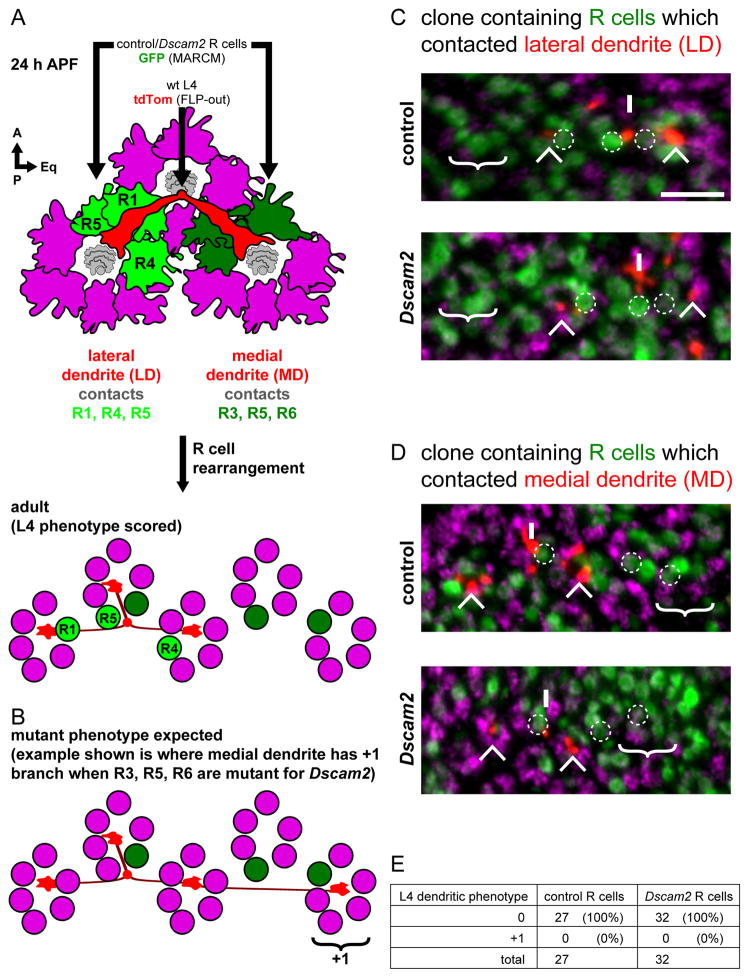
Figure 5. Dscam2 directs L4 dendritic targeting independent of R cells.
(A) Schematic of L4 neuron dendrites (red) and surrounding R cell growth cones (magenta) at 24 h APF. Each dendrite projects beneath two groups of R cell growth cones; the lateral dendrite (LD) contacts R1, R4 and R5 (light green) while the medial dendrite (MD) contacts R3, R5 and R6 (dark green). After R cell rearrangement (see text), the R cells end up in different cartridges relative to the L4 neuron in the adult. This is the stage at which we could score for L4 phenotypes (see C and D).
(B) Schematic showing the L4 dendritic phenotype expected should Dscam2 in R cells be required for targeting. Here the medial dendrite produces a +1 branch after having contacted R3, R5 and R6 which are mutant from Dscam2.
(C, D) DL-MARCM was used to label L4 neurons (red; myr-tdTom) in the background of MARCM generated clones of R cells (green; GFP) (see Figure S6). (C) Example of R cells contacted by the lateral dendrite (i.e. R1, R4 and R5; dotted circles) were in a control (top panel) or Dscam2 (bottom panel) clone. In both control and Dscam2 experiments, no ectopic branching was observed; the cartridge where an ectopic branch would be expected is devoid of any L4 dendrites (‘no +1’). (D) Clones of R cells that contacted the medial dendrite (i.e. R3, R5 and R6; dotted circles) are identified in using the same experimental approach as in (C). Again, in no case was an ectopic branch observed in the expected cartridge (‘no +1’). L4 axons (Ax), lateral (LD) and medial dendrites (MD) are indicated. Processes from non-relevant L4s are also indicated (*). Anti-Hiw (magenta) labels all cartridges. Scale bar, 5 μm. (E) Quantification of L4 dendrites having a 0 or +1 phenotype when in the background of control or Dscam2 mutant R cells.