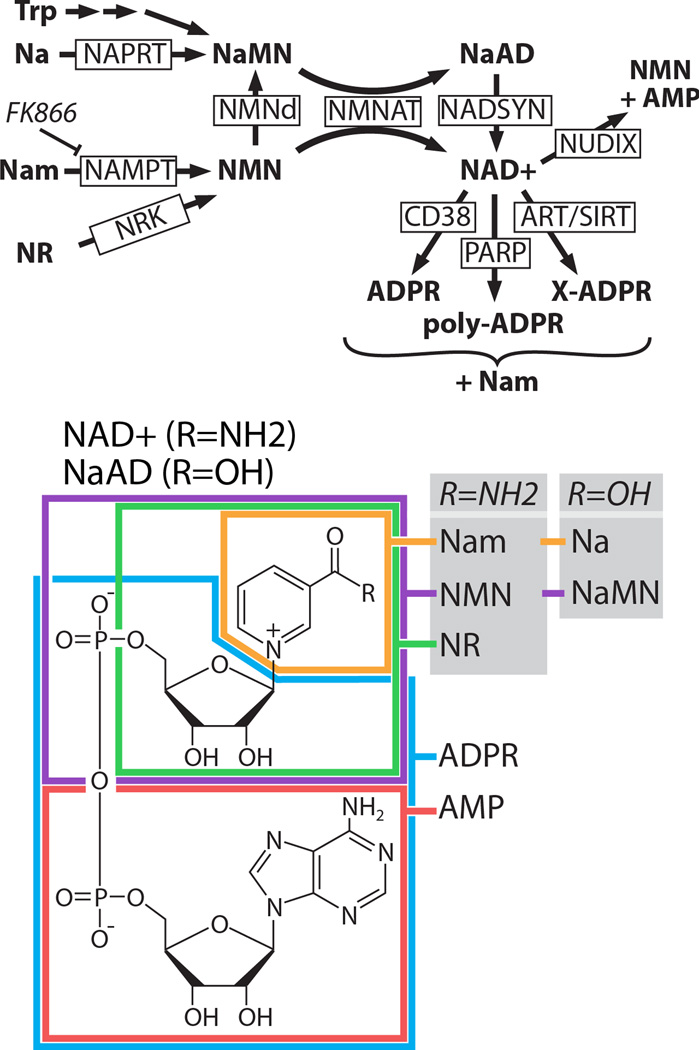
Figure 2.
Pathways of NAD+ synthesis and breakdown. TOP: NAD+ is synthesized from nicotinamide (Nam), nicotinic acid (Na), nicotinamide riboside (NR), or tryptophan (Trp). All synthetic pathways require NMNAT. Nicotinic acid mononucleotide (NaMN) can be synthesized from nicotinamide mononucleotide (NMN) in some bacteria by the enzyme NMN deaminase (NMNd), which has no mammalian ortholog. NAD+ is broken down by multiple classes of enzymes including the glycohydrolase CD38 (Aksoy et al., 2006), poly-ADP-ribose polymerases (PARPs), NUDIX phosphohydrolases (McLennan, 2006), ADP ribosyltransferases (ARTs), and sirtuins (SIRTs). PARPs create polymers of ADPR that are usually attached to a protein substrate. ARTs transfer adenosine diphosphate ribose (ADPR) from NAD+ to an acceptor molecule (X) such as a protein. SIRTs transfer an O-acetyl group from a protein substrate to the ADPR moiety of NAD+ to yield O-acetyl-ADPR and Nam. NaMN=nicotinic acid mononucleotide; NaAD=nicotinic acid adenine dinucleotide; NMN=nicotinamide mononucleotide; NAD+=nicotinamide adenine dinucleotide; AMP=adenosine monophosphate; NAPRT=nicotinic acid phosphoribosyltransferase, NMNAT=nicotinamide mononucleotide adenyltransferase, NADSYN=NAD synthetase, NAMPT=nicotinamide phosphoribosyltransferase; NRK=nicotinamide riboside kinase, NUDIX=nucleoside diphosphate moiety linked X. BOTTOM: Structure of NAD+ with substrate moieties approximately outlined. For a more detailed overview of this pathway, please see these reviews: (Belenky et al., 2007; Chiarugi et al., 2012).