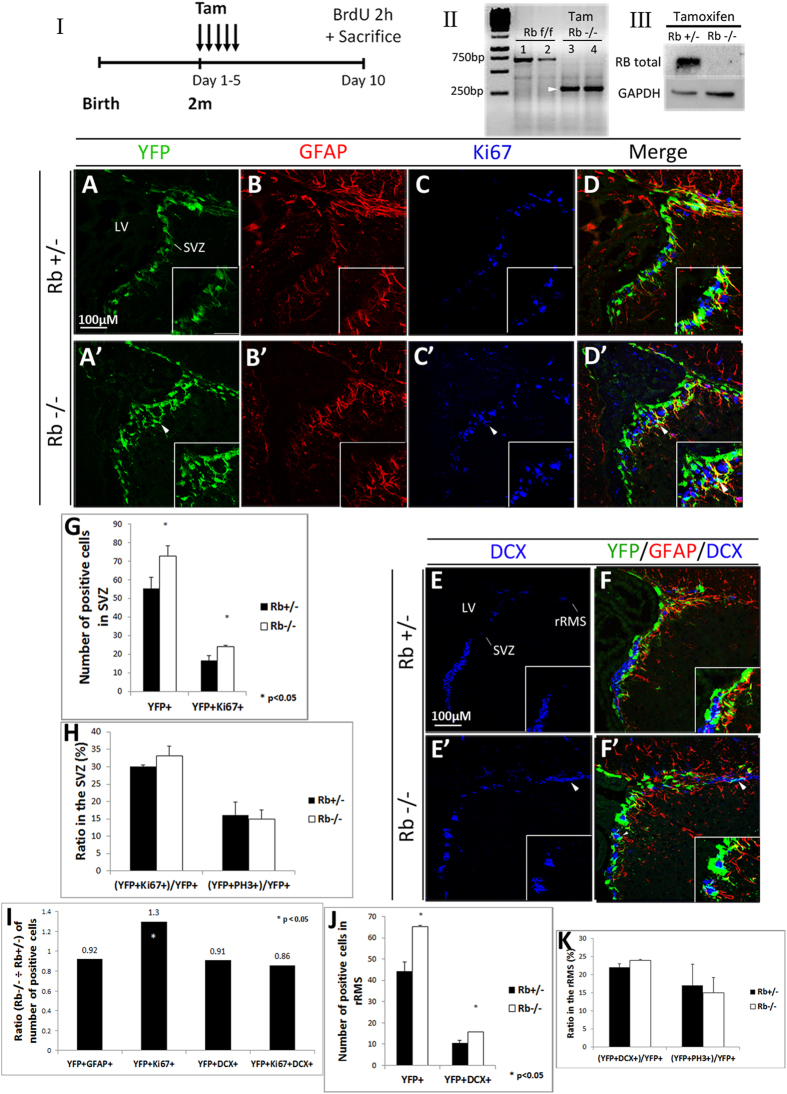
Figure 1. Induced temporal deletion of Rb in aNSCs and progenitors in the SVZ.
(I) Experimental design for the temporal deletion of Rb: animals received 5 tamoxifen (Tam) injections (1 injection per day) at 2 months (2m) of age and a single BrdU injection 2 h prior to sacrifice on day 10. (II) PCR analysis of gDNA extracted from Rbflox/flox neurospheres (1, 2) and Rb−/− green fluorescent neurospheres (3, 4): arrow point to a 320 bp band corresponding to the Rb cleaved allele. (III) Western blot analysis of protein extracts from Rb+/− and Rb−/− green fluorescent neurospheres showing loss of Rb protein. (A–D′) Triple immunohistochemistry performed on sagittal sections against YFP, GFAP and Ki67 in the SVZ in Rb+/− (A–D) and Rb−/− (A′–D′) mice (n = 3Ct and 3Mut). (E–F′) Triple staining with anti-YFP, anti-GFAP and anti-DCX in Rb+/− (E,F) and Rb−/− (E′,F′) mice (n = 3Ct and 3Mut). Arrowheads in (A′–F′) depict increased numbers of stained cells in Rb−/− compared with Rb+/− mice. Insets in (A–F′) are higher magnifications of a selected region in the medial SVZ. (G,H) quantifications of YFP+ and YFP+Ki67+ cells in the SVZ and the ratios of double positive cells over total YFP+ cells, respectively. (I) shows the ratios (Mut ÷ Ct) of YFP+ cells co-labeled with distinct cell markers. (J,K) quantifications of YFP+ and YFP+DCX+ cells in the rRMS and the ratios of double positive cells over total YFP+ cells, respectively. Error bars represent SD of measurements from at least n = 3 per genotype and asterisks indicate a statistically significant difference between genotypes using independent samples t-tests. Scale bar = 100 uM. f; flox, LV; lateral ventricle, rRMS; rostral part of the rostral migratory stream, SVZ; subventricular zone.