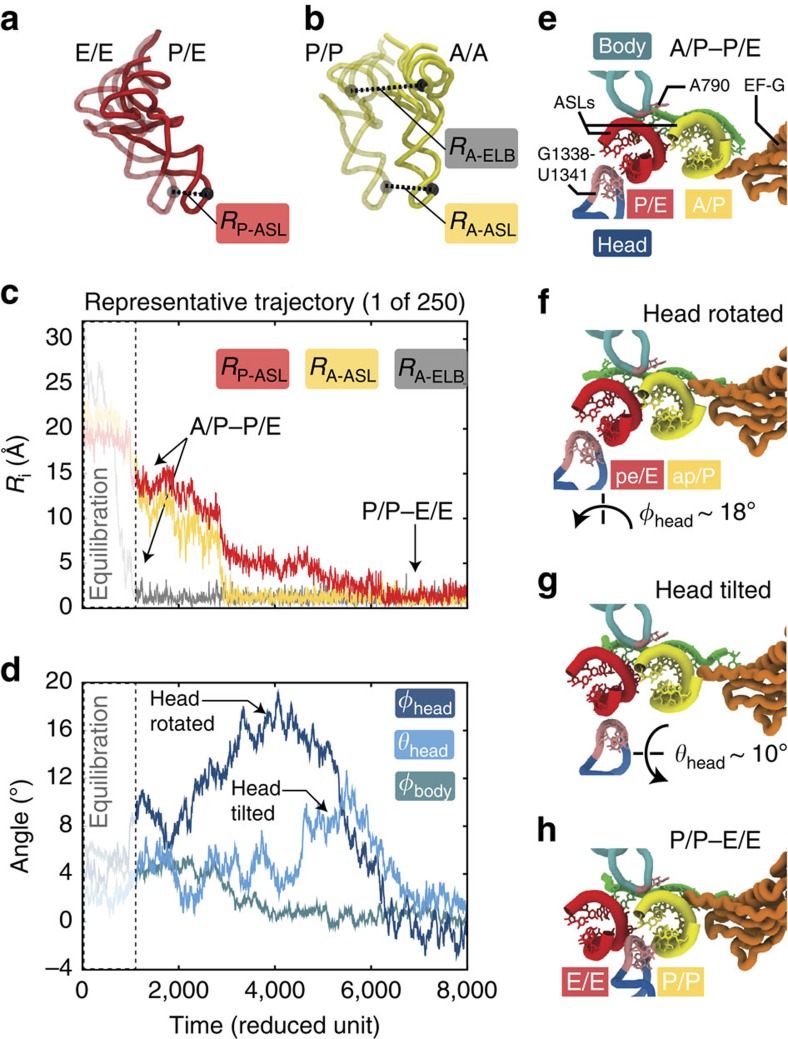
Figure 2. Complex 30S translocation dynamics emerge from a simple energetic model.
(a,b) Multiple coordinates are used to describe tRNA movement. RP−ASL and RA−ASL are distances of the anticodon stem-loops (ASLs) of the P and A site tRNAs (red and yellow) to their E/E and P/P positions. These ASL distances were calculated after 30S body alignment of the simulated trajectory to the POST conformation (see Supplementary Information for details). RA−ELB is the distance of the A-tRNA elbow to its location when in the P/P conformation. Elbow distance is determined after 23S alignment of the trajectory to the POST conformation. In a and b, the P/E and A/A tRNA conformations (initial configuration in the simulation) are shown as opaque. The endpoint E/E and P/P conformations are in ghost representation. (c,d) Representative time traces for a single simulated event (1 of 250) shows large-scale movements of the tRNAs (20–30 Å, c) and transient subunit rotations in the ribosome (d). In all 250 simulations, formation of the A/P–P/E conformation, where RA−ELB (grey line) decreases from ∼30 to ∼0 Å, precedes displacement of the ASLs (red and yellow lines). In c and d, the dashed white-faded boxes outline the initial equilibration period, where the A-site tRNA elbow relaxes to the 50S P site and adopts an A/P conformation. (e–h) Structural snapshots of the mRNA binding track (from the perspective of the 50S subunit) during translocation illustrate the tRNA ASL positions, relative to the 30S body and head. During the initial equilibration period, the system adopts an A/P–P/E conformation (e). During translocation, the 30S head rotates (f), which is followed by back rotation and tilting of the head (g). Translocation is completed when the classical P/P–E/E conformation (h) is reached.