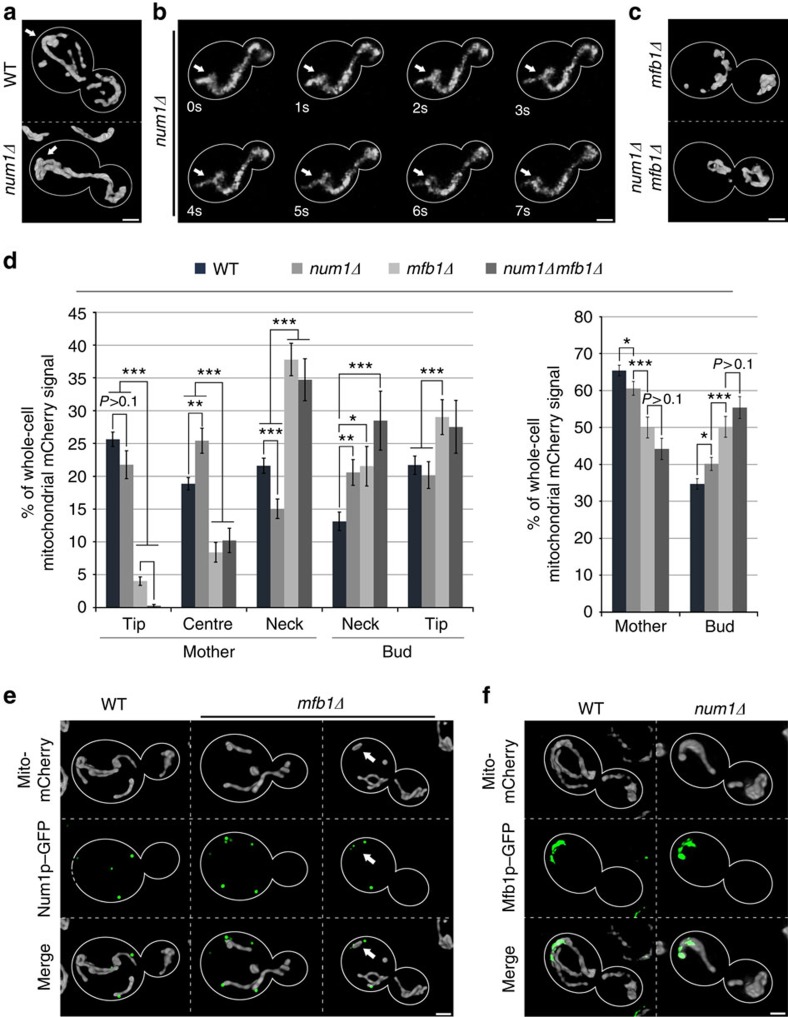
Figure 2. Mfb1p localizes to the mother cell tip and is required for Num1p-independent anchorage of mitochondria at that site.
(a–c) Cells expressing Cit1p-mCherry were grown to mid-log phase and imaged by fluorescence microscopy. Cell outlines are shown in white. (a) Representative 3D renderings of mitochondria in wild-type (WT) and num1Δ cells. Arrows point to mitochondria that accumulate in the mother cell tip in both genotypes. (b) Frames from a representative time-lapse series showing a mitochondrial ‘springback' event at the mother tip of a num1Δ cell. Arrows mark the initial position of a mitochondrion that undergoes anterograde movement (t=1–6 s) and retraction (t=6–7 s). (c) Representative 3D renderings of mitochondria in in mfb1Δ and num1Δ mfb1Δ cells. (d) Quantification of the relative distribution of mitochondrial Cit1p-mCherry in WT, num1Δ, mfb1Δ and num1Δ mfb1Δ cells in five zones as defined in Fig. 1b. Error bars show the s.e.m. (n>40). Data are representative of three independent experiments. (e,f) Cells expressing Cit1p-mCherry and endogenously tagged Num1p-GFP or Mfb1p-GFP fusion proteins were grown to mid-log phase and imaged by fluorescence microscopy. (e) Representative 3D renderings of mitochondria and Num1p-GFP in WT and mfb1Δ cells. White arrows indicate residual Num1p-dependent mitochondrial retention at the mother tip in the absence of Mfb1p. (f) Representative 3D renderings of mitochondria and Mfb1p-GFP in WT and num1Δ cells. Scale bars, 1 μm. Statistical significance was determined using Student's t-test. *P<0.05, **P<0.01 and ***P<0.005.