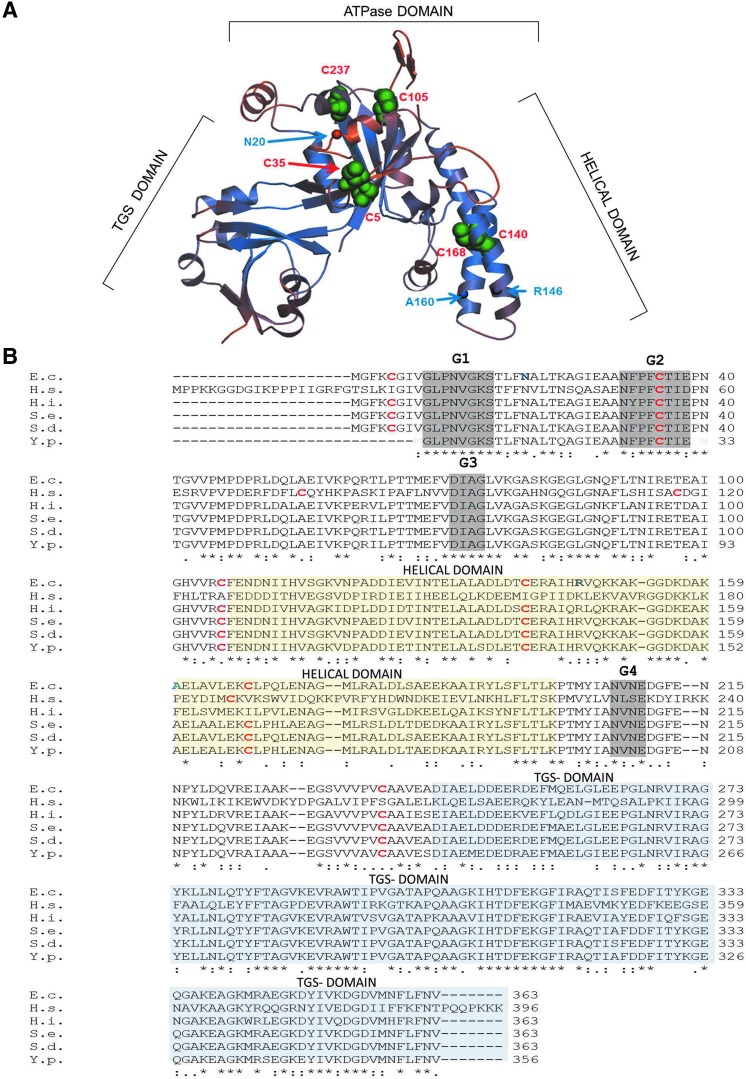
FIG. 1.
Position and sequence conservation of cysteine residues in YchF/Ola1. (A) The crystal structure of the YchF homologue of Haemophilus influenzae (1JAL) was used to model the probable position of the six cysteine residues in Escherichia coli YchF using the Swiss Model (http://swissmodel.expasy.org/interactive/kFyFA3/models). Cysteine residues are labeled with green spheres and the amino acid position based on the E. coli numbering is indicated in red. In addition, the residues where the UV-induced cross-linker pBpa was incorporated are indicated by circles and the amino acid position based on the E. coli numbering is indicated in blue. (B) A sequence alignment of YchF homologues from different enterobacteria was generated by using the Clustal 2.0 software and compared with the H. influenzae YchF and Homo sapiens Ola1 sequence. Cysteine residues are shown in red and bold and the positions where pBpa was incorporated in the E. coli YchF are shown in blue and bold. The G1-G4 motifs of the nucleotide-binding site are boxed in gray, the helical domain is boxed in yellow, and the TGS domain in blue. E.c., Escherichia coli; H.s., Homo sapiens; H.i., Haemophilus influenzae; S.e., Salmonella enterica; Y.p., Yersinia pestis; S.d., Shigella dysentericae. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars