Figure 1.
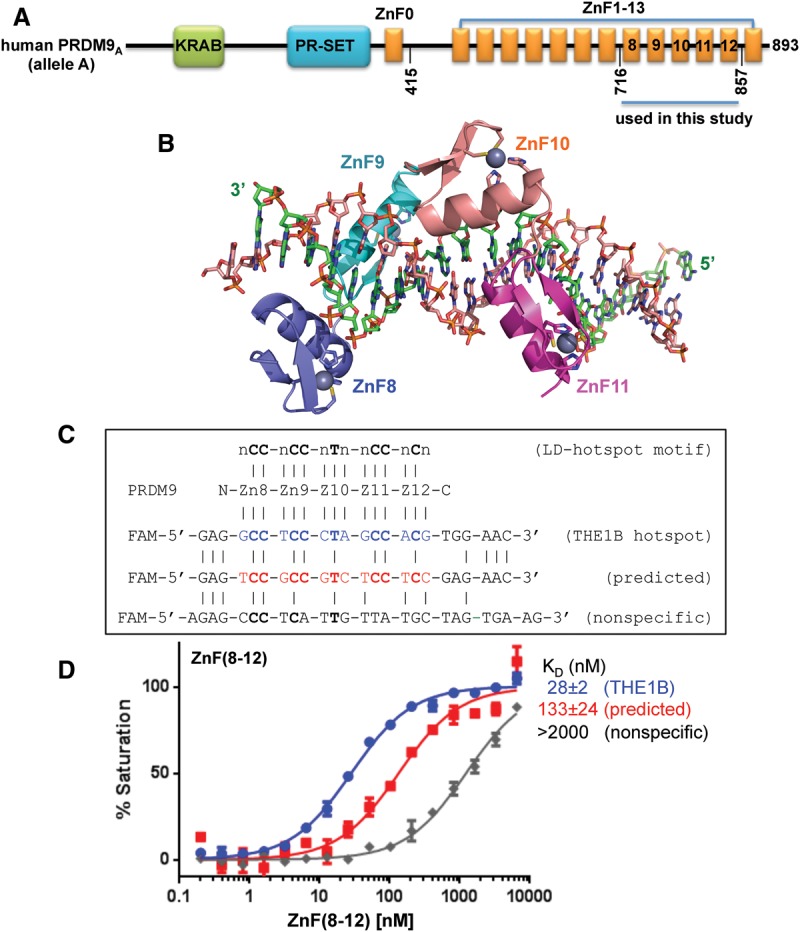
PRDM9A DNA binding. (A) Domain organization of the hPRDM9 protein (accession no. Q9NQV7). (B) Crystal structure of hPRDM9A (hPRDM9, allele A) ZnF8–12 in complex with the THE1B recombination hot spot sequence. ZnF8–11 (color-coded blue [ZnF8], cyan [ZnF9], orange [ZnF10], and magenta [ZnF11]) are shown in cartoon representation. ZnF12 was not visible in the structure. The helix of each finger forms specific H bonds in the major groove mainly with bases in the DNA strand (green) that is complementary to the consensus sequence (orange). The ZnF array, oriented N to C from left to right, interacts with this DNA strand oriented 3′ to 5′. (C) DNA sequences of THE1B hot spot (Myers et al. 2008), a predicted sequence (Baudat et al. 2010), and an arbitrary negative control that partially overlapped the consensus. The top line indicates the LD-hot spot motif (Myers et al. 2008), which was derived from recombination hot spots that are intrinsically sex- and population-averaged (Pratto et al. 2014). (D) PRDM9A ZnF(8–12) binds the THE1B hot spot sequence.
