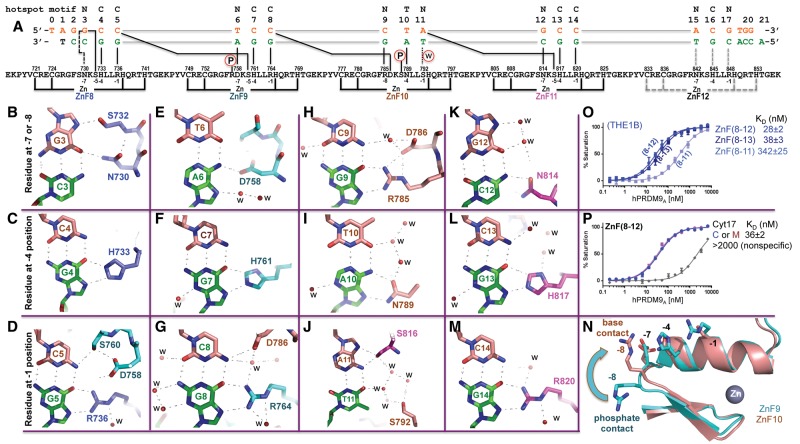
Figure 2.
PRDM9A binds the THE1B hot spot. (A) Schematic representation of the ZnF8–12 DNA-binding domain. The top line indicates the LD-hot spot motif. The second line indicates the base pair positions (1–20). The third and the fourth lines are the sequence of the oligos used for this study, shown with the top strand (orange) oriented left to right from 5′ to 3′, matching the 13-mer LD-hot spot motif sequence. The complementary G-rich strand (green) has the base-specific interactions with each ZnF. Two cysteine and two histidine residues (C2H2) in each finger are responsible for Zn2+ ligand binding (bottom connecting lines). Amino acids at positions −1, −4, and −7 (or −8) relative to the first histidine interact specifically with the DNA bases shown below. (B,E,H,K) DNA base interactions involve a residue at position −7 or −8 of each ZnF. (C,F,I,L) DNA base interactions involve a residue at position −4 of each ZnF. (D,G,J,M) DNA base interactions involve a residue at position −1 of each ZnF. (N) Superimposed ZnF9 (cyan) and ZnF10 (orange). Arg757 of ZnF9 at the −8 position makes a DNA–phosphate interaction, while Arg785 of ZnF10 at the −8 position makes a DNA base interaction. (O) Comparison of ZnF8–11, ZnF8–12, and ZnF8–13 with oligos containing THE1B hot spot sequence. (P) Comparison of ZnF8–12 with oligos containing unmodified C or 5-methyl-cytosine (M) at the G:C base pair of position 17.