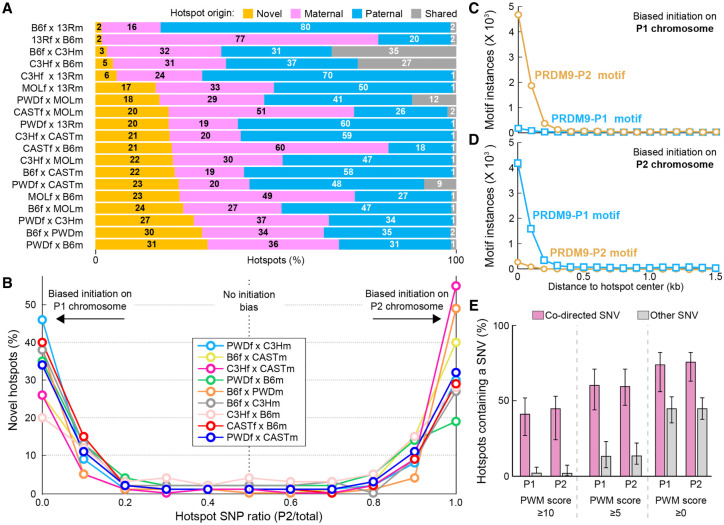
Figure 2.
Novel hot spots in F1 hybrid mice. (A) In F1 hybrid mice, up to 31% of hot spots occur at sites that are not used in parental mice (novel hot spots; orange). (B) A majority of novel hot spots exhibit strongly biased DSB formation on one or the other parental chromosome. To generalize findings across multiple strains, we refer to parental chromosomes as P1 (maternal) and P2 (paternal). Initiation biases were determined by examining SNPs between parental genomes. Hot spots were binned in deciles by the fraction of ssDNA-de-rived sequencing reads overlapping SNP loci that contained P2-derived SNPs. (C) P2 PRDM9 motifs are enriched at novel hot spots where DSBs exhibit an initiation bias on the P1 chromosome. (D) P1 PRDM9 motifs are enriched at novel hot spots where DSBs exhibit an initiation bias on the P2 chromosome. (E) A large percentage of hot spots with biased initiation is explained by SNVs between parental genomes. DSB hot spots with an initiation bias were split by initiation bias (P1,P2). The proportion of DSB hot spots that contain a codirected SNV (pink) in the central 500 base pairs (bp) was calculated. Other, noncodirected SNVs were also examined to give an estimate of the expected background variation rate. Bar height represents the average value across all nine F1 strains for which SNV data are available for both parental strains. Error bars represent the maximum and minimum values across all F1 strains. Data for progressively more lenient motif alignment score thresholds are shown from the left to the right panels. The PWM score threshold is surpassed when either parental chromosome harbors a motif that exceeds the score threshold. There is a large excess of codirected SNVs at hot spots that exhibit biased initiation.