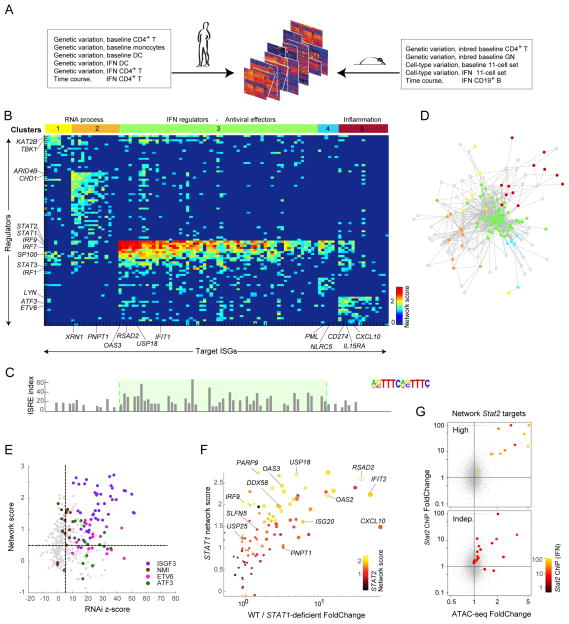
Fig. 5. Trans-species regulatory network inference.
(A) Datasets used for network inference. (B) Heatmap of inferred regulatory scores (sparsified at FDR<0.01) connecting regulators in rows (kinases, phosphatases, TFs) and targets (columns), color-coded to strength of predicted association. Regulators and targets biclustered into 5 clusters, color-coded at top. (C) FIMO scores in TSS of target ISGs for ISRE. (D) Network-based representation of regulator-target links, color-coded by cluster membership (per B); only showing links passing Bonferroni p<10−6. (E) Each testable regulator-target link shown as a dot, placed by its z-score from regulator RNAi knockdown ((Amit et al., 2009)) and network score. Predicted links for ISGF3, NMI, ETV6, and ATF3 in color. (F) Correspondence between predicted STAT1 regulator score for each ISG and expression Foldchange in fibroblasts from a STAT1-deficient patient. Predicted STAT2 regulatory strength for these ISGs color-coded. Dot size shows magnitude of response in wild-type condition. See Fig S5 and Tab.s S3B-G. (G) Relation between IFN-induced changes in ATAC peak intensity and Stat2 binding for ISGs with top or bottom 10% Stat1/2 network scores (color-coded for Stat2 signal intensity).