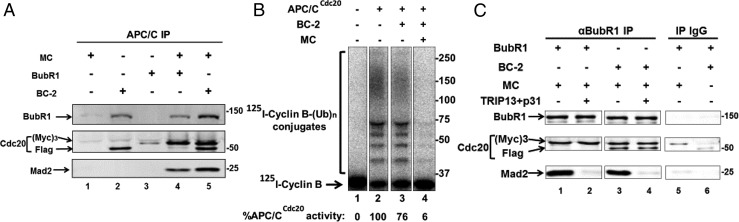
Fig. 3.
Mad2-Cdc20 can combine with BC-2 to form MCC with an additional Cdc20 (MCC-1–2). (A) Formation of MCC-1–2 and its binding to APC/C. Incubation conditions were as described in Fig. 2A. The indicated subcomplexes and BubR1 were added at 150 nM. Reaction products were added to APC/C bound to anti-Cdc27 beads. Following further incubation for 60 min at 23 °C, proteins bound to APC/C were isolated by precipitation of anti-Cdc27 beads, followed by washing and APC/C elution with Cdc27 peptide as described in Materials and Methods. Subsequently, supernatants were immunoblotted for the indicated proteins. Numbers on the right indicate the electrophoretic migration of the marker proteins (kDa). (B) MCC-1–2 is an inhibitor of APC/CCdc20. BC-2 was incubated with or without MC as indicated. Incubation conditions were as described in Fig. 2A, and the final concentration of the specified subcomplexes was 100 nM. Subsequently, the products were added to APC/CCdc20, and following incubation their effects on cyclin B–ubiquitin ligation activity were determined as described in Fig. 2B. The results indicated at the bottom are expressed as the percentage of I125-cyclin B-(Ub)n conjugates formed relative to APC/CCdc20 without additions. Numbers on the right indicate the migration position of marker proteins (kDa). (C) MCC-1–2 is disassembled by the joint action of TRIP13 and p31comet. Different combinations of Mad2-(Myc)3-Cdc20 (MC) with either BubR1 or BubR1-Flag-Cdc20 (BC-2) were incubated at conditions described in Fig. 2A and then were precipitated with either anti-BubR1 or IgG beads, as indicated. Subsequently, beads were washed three times with Buffer A and then were incubated with TRIP13 and p31comet, where indicated, under conditions similar to those described in SI Materials and Methods for BC-1 formation. Sham treatment was under similar conditions but without TRIP13 and p31comet. Finally, the beads were washed, and samples were subjected to immunoblotting for the indicated proteins. Numbers on the right side indicate the migration position of marker proteins (kDa).