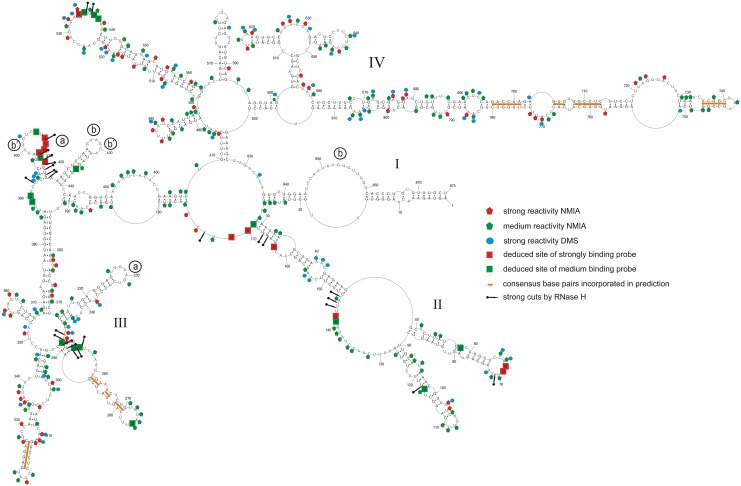
Fig 1. Self-folding of vRNA8 predicted by RNAstructure 5.3 using as constraints: strong reactivity of DMS; consensus base pairs from sequence and structure analysis (orange bars); SHAPE reactivities converted to pseudo- free energies.
Additionally there are marked results from microarray mapping in buffer A (300 mM NaCl, 5 mM MgCl2, 50 mM HEPES, pH 7.5) at 37°C and also from RNase H cleavage in the same buffer and temperature. Binding sites of probes are denoted by the middle nucleotide of the five nucleotides complementary in the RNA. Possible regions of tertiary interactions are marked by letters in open circles—the same letter marks one interaction (see text). Regions with no read-out by chemical mapping are: 807–875 (NMIA) and 835–875 (DMS). The numbering of vRNA8 is from its 5’ end. The template for the AUG start codon is nucleotides 849–847.