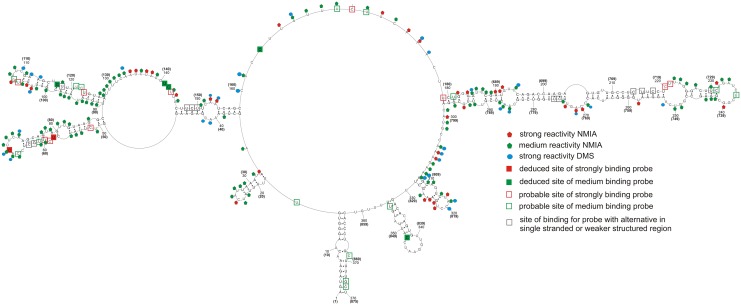
Fig 5. Self-folding of mini-vRNA8 predicted by RNAstructure 5.3 using as constraints: strong reactivity of DMS and SHAPE reactivities converted to pseudo-energy.
Additionally there are marked results from microarray mapping in buffer A (300 mM NaCl, 5 mM MgCl2, 50 mM HEPES, pH 7.5) at 37°C. All symbols are the same as in Figs 1 and 4. The regions without readout of chemical mapping results are: 327–376 (826–875) (NMIA), and 322–376 (821–875) (DMS). Numbering of mini-vRNA8 is from its 5’ end and numbers in parenthesis correspond to respective nucleotides in vRNA8. Nucleotides 183–187, 5’GGAUC, were introduced for cloning (see Materials and methods). Nucleotides 1–182 and 188–376 correspond to wild type. Efficient packaging of a segment 8 encoding only GFP protein required nucleotides 1–177 and 198–376 (mini-vRNA8 nomenclature) [45].