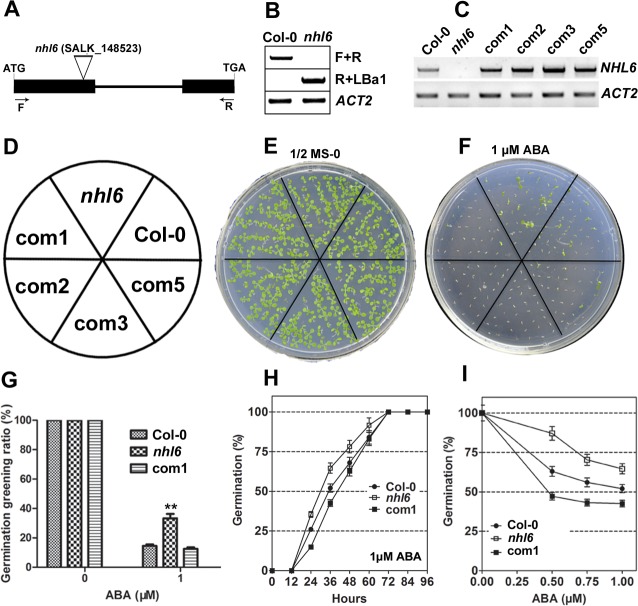
Fig 2. Molecular identification and ABA insensitivity of nhl6 mutant.
(A) Schematic diagram of T-DNA insertion in nhl6 mutant. Exons and intron are depicted to scale by boxes and line, respectively. The coding region of NHL6 is shown as black boxes. The position of T-DNA is indicated by triangle. (B) PCR analysis to confirm the disruption of NHL6 gene by T-DNA insertion. The positions of primers F and R are indicated in (A). ACTIN2 was used as a native control. (C) RT-PCR analyses of NHL6 gene expression in wild-type Col-0, homozygous nhl6 knockout mutant, and the complemented ProNHL6::NHL6/nhl6 transgenic lines 1, 2, 3 and 5 (com1, 2, 3, 5). Total RNA was isolated from the rosette leaves of four-week-old plants. Expression of ACTIN2 (ACT2) was analyzed as an internal control. (D-F) Seeds of Col-0, nhl6 and the complemented ProNHL6::NHL6/nhl6 transgenic lines 1, 2, 3 and 5 were sowed on MS medium supplemented with or without 1μM ABA. Photographs were taken after one week. (G) Statistical analyses of seed germination greening ratio in Col-0, nhl6 and ProNHL6::NHL6/nhl6 transgenic line 1 (com1). After the vernalization, seeds on MS medium plates were transferred to a growth chamber and cultured for 7 days. Values are means ±SE, n = 3 independent experiments. (H) Germination time course (hours after incubation at 22°C) of Col-0, nhl6 and ProNHL6::NHL6/nhl6 transgenic line 1 on MS medium supplemented with 1 μM ABA. (I) Germination rates of Col-0, nhl6 and ProNHL6::NHL6/nhl6 transgenic line 1 after 2 days at 22°C in the presence of different concentrations of ABA. Col-0, closed circles; nhl6, open squares; com1, closed squares. Results are presented as average values and standard errors from three experiments. At each time, at least 100 seeds were counted.