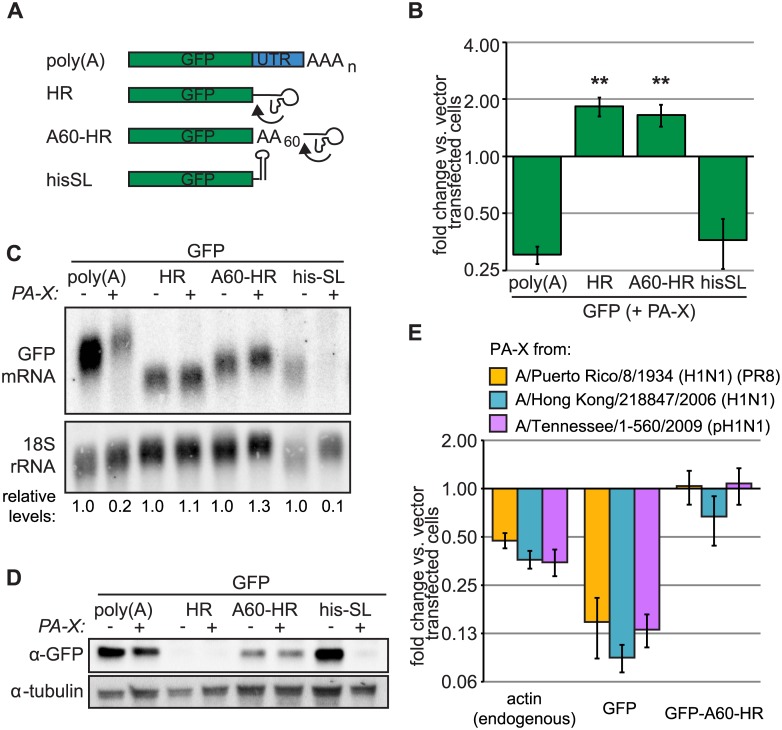
Fig 6. Canonical processing of the 3’ end of Pol II transcripts confers sensitivity to PA-X.
(A) Diagram of the GFP reporter constructs used in the figure, which bear different 3’ mRNA ends: the canonical poly(A) tail added by cellular processing machinery (poly(A)), a hammerhead ribozyme (HR), the hammerhead ribozyme proceeded by a 60-nt templated poly(A) stretch (A60-HR), or the histone stem-loop structure (hisSL). The arrows indicate where the HR self-cleaves the RNA. (B-D) HEK 293Ts were transfected with indicated Pol-II driven GFP reporters and either PA-X or empty vector. Total cellular RNA was extracted and GFP mRNA levels were measured by RT-qPCR (B) or northern blotting (C). GFP expression normalized by 18S rRNA abundance is plotted (B) or reported (C, relative levels) as fold change in PA-X-expressing vs. vector-transfected cells. (D) Levels of GFP for experiments in panel B were assessed by western blot, using tubulin as a loading control. A representative image is shown. ** = p value (Student’s t-test) < 0.01 for fold change of indicated reporter vs. fold change of poly(A) GFP. (E) HEK 293Ts were transfected with indicated Pol-II driven GFP reporters and PA-X from three different IAV strains or empty vector. Total cellular RNA was extracted and GFP or actin mRNA levels were measured by RT-qPCR. GFP expression normalized by 18S rRNA abundance is plotted as fold change in PA-X-expressing vs. vector-transfected cells. Error bars = s.e.m.