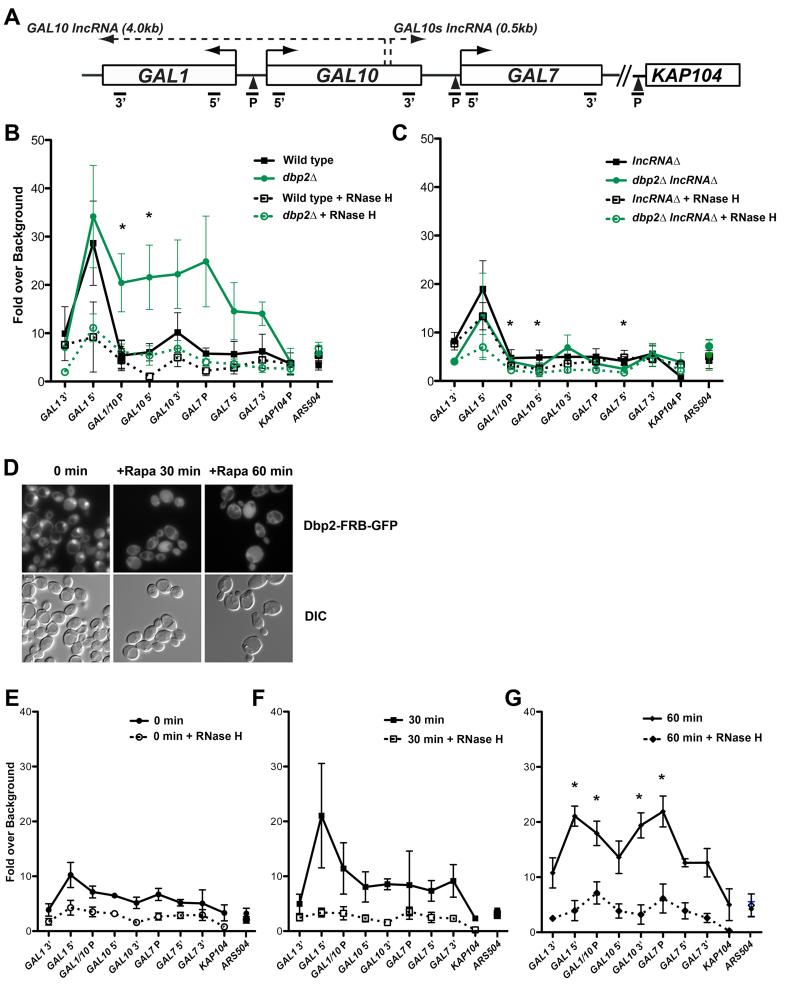
Figure 3. Loss of Dbp2 promotes accumulation of GAL lncRNA R-loops across the GAL cluster.
(A) Schematic of the GAL gene cluster showing coding regions, GAL10 and GAL10s lncRNA loci, and qPCR primer-probe positions. (B-C) DNA-RNA-immunoprecipitation (DRIP) of wild type and dbp2Δ (B) or lncRNAΔ and dbp2Δ lncRNAΔ (C) cells grown under transcriptionally repressive (+glucose) conditions. DRIP was conducted using the S9.6 antibody (Boguslawski et al., 1986; El Hage et al., 2010; Hu et al., 2006) or beads alone (no antibody control) followed by qPCR using the indicated primer-probe sets (A) or the non-protein coding region ARS504, which lacks both RNase H-dependent R-loops (Chan et al., 2014) and Dbp2-dependent transcripts (Beck et al., 2014). R-loop levels were determined across 5 biological replicates as the increase in signal of above the no antibody control presented as the mean and S.E.M. * = p < 0.05. (D) Rapamycin-dependent cytoplasmic localization of Dbp2-GFP via anchor away. Dbp2-FRB-GFP was visualized by fluorescent microscopy 0 min (-Rapa), 30 min or 60 min after treatment with 10μg/mL rapamycin. (E-G) Cytoplasmic relocalization of Dbp2 promotes time-dependent accumulation and spreading of R-loops across the GAL cluster. DRIP was conducted as in B with cells following 0, 30 or 60 min rapamycin treatment. * = p < 0.05 with respect to 0 min.