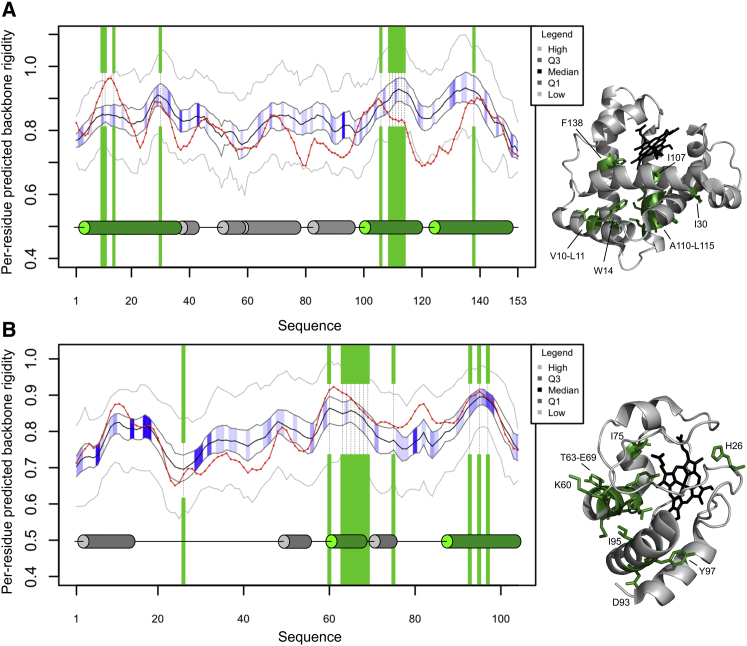
Figure 5.
The structural, dynamics and evolutionary properties of sperm whale apo-Mb (A) and horse ferricytochrome c (B) are shown as a function of their residue positions on the left, whereas the corresponding three-dimensional structures are on the right (PDB: 1MBC and 1HRC, respectively; structures are only available for the proteins together with their heme cofactors (black), and for cyt c the wild-type protein is depicted). In both panels, early folding residues are marked with green shading on the graphs and with green stick representations within the three-dimensional structures, with their residue positions and types indicated. A red line depicts the per-residue DynaMine-predicted backbone rigidity of the protein. The medians of predicted values in the corresponding HHBLITS_lowSeqId alignment columns are shown as a black line, whereas their first and third quartiles of the distribution are marked in dark gray, their minima and maxima with lighter gray. The blue shading between the dark gray quartile lines represents the sequence entropy for each alignment position, with darker blue indicating lower entropy (high evolutionary conservation). The secondary structure elements assigned by the POLYVIEW server are also provided, with early folding helices shown as green cylinders and others as gray cylinders. To see this figure in color, go online.