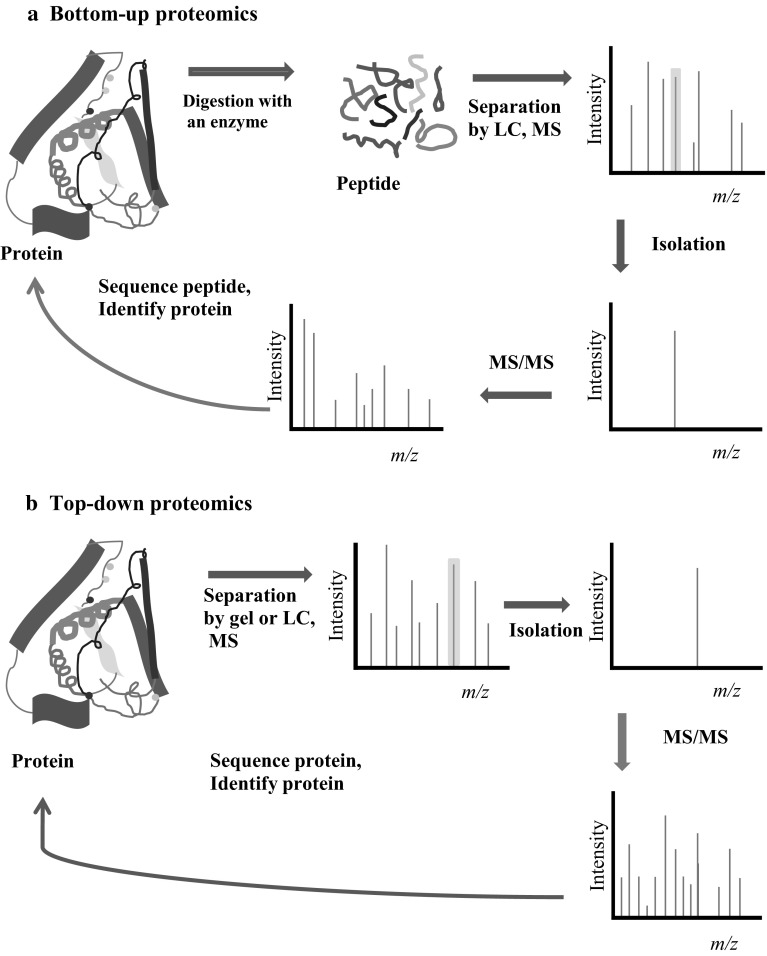
Fig. 1.
Principal differences between top-down (a) and bottom-up (b) proteomics. a In bottom-up proteomics, protein extracts are separated by 2-dimensional gel electrophoresis (2-DE) and excised gels are digested with chemical tags or separated by liquid chromatography (LC) after protein extracts are digested with chemical tags. Labeled peptides are analyzed and isolated by mass spectrometry (MS) and fragmented by tandem mass spectrometry (MS/MS) to identify the protein from the database. Consequently, hundreds of proteins can be identified and quantified with significant confidences but the sequence coverage of proteins is far from complete sequence coverage. (b) In top-down proteomics, a complex sample, such as a tissue sample, is separated by 2-DE or LC and analyzed directly by MS/MS without digestion. Thereby, this strategy can analyze the protein’s full sequence coverage