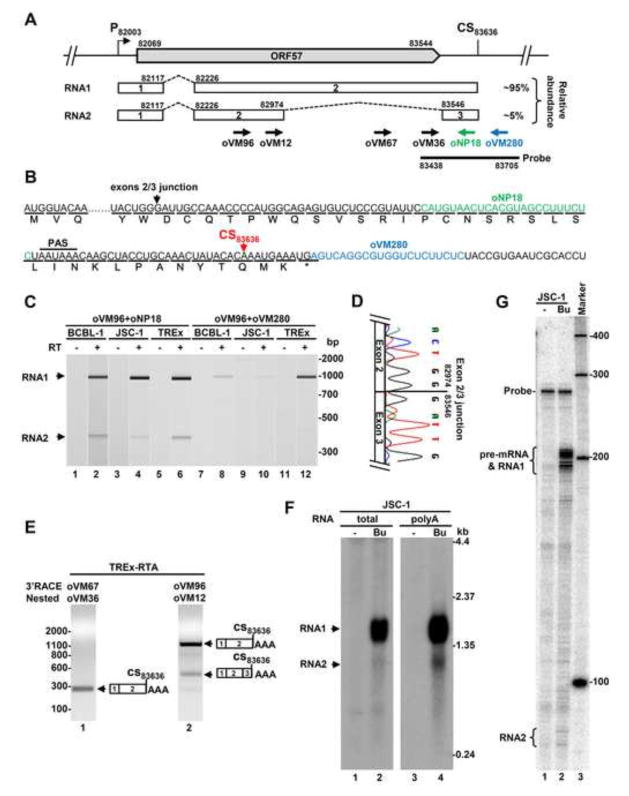
Fig. 1.
KSHV ORF57 gene encodes two isoforms of alternatively spliced RNA. (A) Diagram of the KSHV ORF57 locus in the virus genome showing the ORF57 coding region (big grey arrow), the transcriptional starting site (P) at nt 82003 and the polyadenylation cleavage site (CS) at nt 83636. The alternatively spliced RNA transcripts (RNA1 and RNA2) are shown below. The numbered boxes represent exons separated by introns (dashed lines). On the right is the expression level of individual ORF57 transcripts determined from the Northern blots shown in Fig. 1F determined by ImageJ analysis (http://imagej.nih.gov/ij/). Arrows below the RNA2 are primers used in this study, with a thick line for an antisense RNA-probe used for Northern blot and RPA. The nucleotides positions are based on the sequence of the KSHV genome (GenBank Acc. No. U75698.1). (B) Amino acid composition of the hypothetical ORF57A encoded by RNA2 derived from double splicing. Arrows mark the positions of the exon-exon junction and ORF57 polyadenylation cleavage site (CS), respectively. The classical polyadenylation signal (PAS) AAUAAA of ORF57 and reverse primers (colored) upstream or downstream of the CS83636 site are also indicated in this panel. (C) Alternative splicing of ORF57 exon 2 during KSHV lytic infection in three KSHV-infected primary effusion lymphoma cell lines. The viral lytic cycle was induced by 48 h treatment with 1 mM valproic acid (BCBL-1), 3 mM sodium butyrate (JSC-1) or 1 μg/ml of doxycycline (TREx BCBL1-RTA). Two-step RT-PCR was carried out on total RNA by reverse transcription (RT+) with random hexamers followed by 25 cycles of PCR amplification with the indicated gene-specific primers. The size and concentration of obtained PCR products were determined by Bioanalyzer 2100 (Agilent). The reactions without RT (−) were used as a negative control. (D) A chromatograph shows the exon-exon junction of ORF57 intron 2 splicing determined by Sanger sequencing of spliced RT-PCR products. (E) Polyadenylation of ORF57 transcripts at the CS83636. The nested 3′RACE was carried out on total RNA from lytic TREx BCBL1-RTA cells. The primers oVM96 and oVM67 were used for the first and oVM12 and oVM36 for the second (nested) amplification. The resulting products were analyzed by agarose gel electrophoresis. The polyadenylation cleavage site was determined by Sanger DNA sequencing of each nested 3′RACE product. (F and G) Expression profile of ORF57 transcripts in KSHV-infected cells. Total or poly A-selected RNA from JSC-1 cells with latent (−) and lytic (Bu-butyrate treated) infection used for ORF57 Northern blot in our previous publication (Majerciak, Yamanegi et al., 2006) was re-probed by an antisense RNA-probe as described in the panel A (F). Total cell RNA from JSC-1 cells with or without butyrate treatment for lytic infection was also examined by RNase-protection assay (RPA) by the same antisense RNA probe described in the panel A (G).