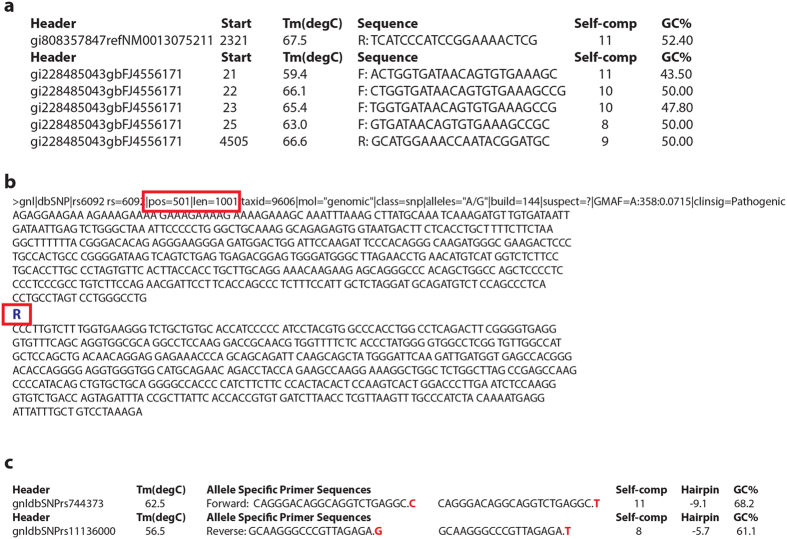
Figure 5. PrimerMapper text based outputs and input SNP sequence requirements.
(a) PrimerMapper generates tab separated value (TSV) based text files that lists the designed primers and their corresponding features from each sequence. (b) In the case of SNP data, PrimerMapper pre-processes all input by collecting the SNP location and SNP type from the rs_fastA formatted input. These features are highlighted by red rectangles. If the SNP sequence data is not remotely retrieved from dbSNP23 , the user can locally upload data, however, the header for each sequence must be unique and contain the SNP location and sequence length as formatted within the red rectangle i.e. “pos = 501|len1001”. Furthermore, the standard SNP sequence format must be adopted with the SNP type e.g. “R” or “Y” etc. inserted within each sequence. (c) PrimerMapper returns a TSV file from SNP data depicting the allele-specific primers for each SNP and the appropriate wildtype and polymorphic primers specific to each SNP, as well as their basic features i.e. sequence header, Tm, Self-complementarity score, ΔG hairpin score, and GC%.