Figure 1. Co-localizations among genomic features.
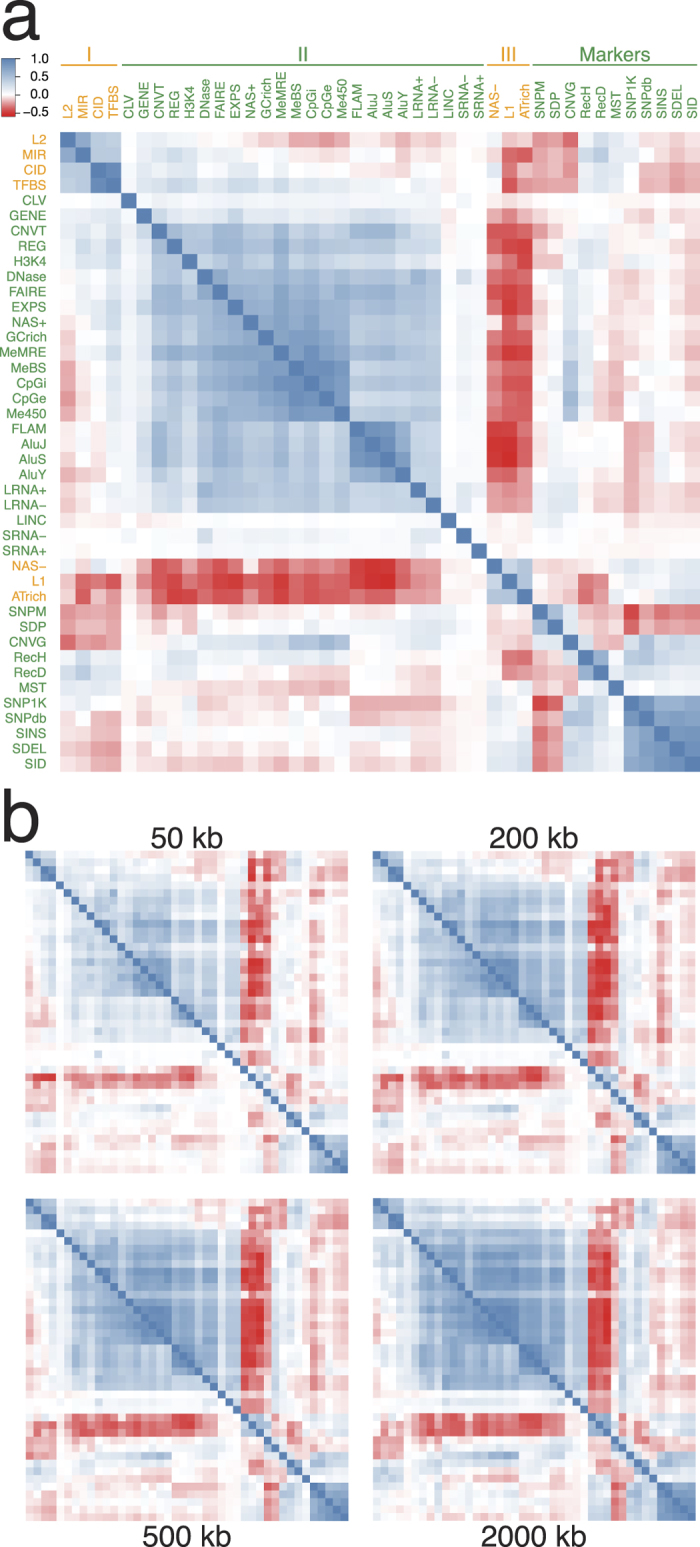
(a) Heat map of pairwise correlation coefficient r among forty-two genomic features in 500-kb non-overlapping windows in the twenty-two autosomes. L2 long interspersed nuclear element 2, MIR mammalian-wide interspersed repeat, CID conserved indel, TFBS conserved transcription factor binding site, CLV breakpoints of variants in ClinVar database, GENE known gene sequences, CNVT somatic CNV breakpoints, REG published DNA regulatory regions, H3K4 histone 3 lysine 4 trimethylation, DNase open chromatin elements and signals, FAIRE formaldehyde-assisted isolation of regulatory element, EXPS gene expression data, NAS+ nuclease accessible sites in CD34+ cells, GCrich GC-rich DNA sequences, MeMRE genome wide methylation data, MeBS CpG methylation, CpGi CpG island, CpGe evolutionarily CpG rich regions, Me450 CpG methylation, FLAM free left Alu monomer, AluJ, AluS and AluY sub-families of short interspersed nuclear element (SINE), LRNA+ long RNA-seq signals of plus strand, LRNA- long RNA-seq signals of minus strand, LINC lincRNA, SRNA- short RNA-seq signals of minus strand, SRNA+ short RNA-seq signals of plus strand, NAS- nuclease accessible sites in CD34- cells, L1 long interspersed nuclear element 1, ATrich AT-rich DNA sequences, SNPM SNPs mapped to multiple locations, SDP segmental duplication, CNVG germline CNV breakpoints, RecH recombination rate from HapMap, RecD sex-averaged recombination rate from deCODE, MST microsatellite, SNP1K single nucleotide polymorphism in 1000 Genomes Project, SNPdb single nucleotide polymorphism in dbSNP, SINS small insertion in dbSNP, SDEL small deletion in dbSNP, SID small indel. Positive and negative co-localizations are expressed in r-values based on the blue and red thermal scales respectively. The partitions of various features into Groups I-III and Markers were based on their nature and co-localization patterns, guided foremost by the strong positive co-localizations. (b) Heat map of pairwise Pearson correlation coefficients (upper triangle) and Spearman correlation coefficients (lower triangle) among genomic features in 50-kb, 200-kb, 500-kb and 2,000-kb windows. Arrangement of different features on x and y axes same as in (a).
