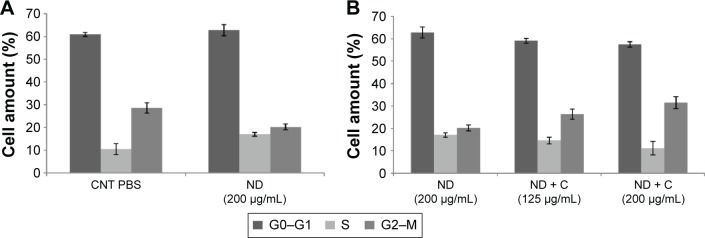
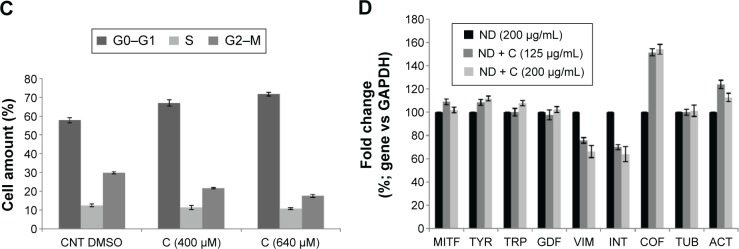
Figure 5.
FACS analysis.
Notes: Cytofluorimetry of B16F10 cells treated for 72 hours with PBS, ND (200 μg/mL), ND + C (125 μg/mL or 200 μg/mL), DMSO, and C (400 μM or 640 μM) is shown (A, B, and C). For each sample, the number of cells detected in the three cell cycle phases (G0–G1, S, and G2–M) is reported in percentage. Gene transcription analysis carried out by real-time PCR was performed after treatment for 72 hours, with ND (200 μg/mL) and ND + C (125 μg/mL or 200 μg/mL) (D). mRNA levels for each gene were first normalized for GAPDH transcript amount and then indicated as percentage of fold change with respect to ND (200 μg/mL) specimen, considered as unit (100%). Data are expressed as mean ± SD and represent the results obtained by performing three independent experiments (P<0.05 vs control for the experiments reported in A–C, and P<0.01 vs control for the experiments in D).
Abbreviations: FACS, fluorescence-activated cell sorting; PBS, phosphate-buffered saline; ND, nanodiamond; C, citropten; DMSO, dimethyl sulfoxide; PCR, polymerase chain reaction; SD, standard deviation; CNT, control; MITF, microphthalmia-associated transcription factor; TYR, tyrosinase; TRP, tyrosinase-related protein; GDF, growth-differentiation factor; VIM, vimentin; INT,ανβ3-integrin; COF, cofilin; TUB, β-tubulin; ACT, β-actin.