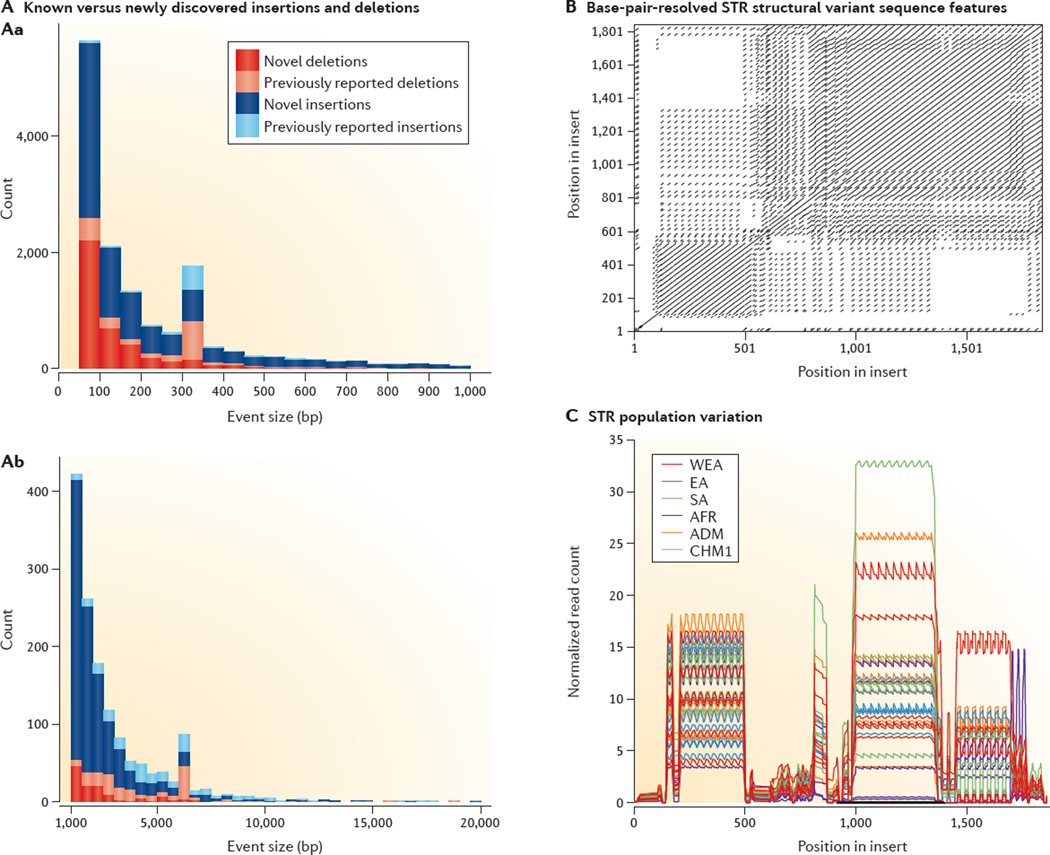
Figure 5. Human genetic variation detected with local assembly of single molecules.
A | Deletions (red and pink) and insertions (dark and light blue) resolved at base-pair resolution in the genome from the CHM1 cell line through local assembly of the single-molecule real-time (SMRT) reads for events less than 1 kb (Aa) and greater than 1 kb (Ab)5. Copy number variants found in previous studies4,57,108 are in lighter shades, with roughly 85% of events being unique to the CHM1 results. B | An example of a 1.7-kb short tandem repeat (STR) insertion event (represented in a self dot plot) not detected by Illumina resequencing of CHM1 but detected and assembled by SMRT reads. C | This STR insertion contains uniquely identifying 30 bp sequences that, once sequence resolved, may be used to genotype the presence of the insertion in genomes sequenced using Illumina technology. Normalized read depth serves as a proxy for estimating variability of STR length and demonstrates that the STR is highly variable in diverse populations (shown for Western Eurasian (WEA), East Asian (EA), South Asian (SA), African (AFR) and admixed (ADM) individuals). Figure adapted from REF. 5, Nature Publishing Group.