FIGURE 2.
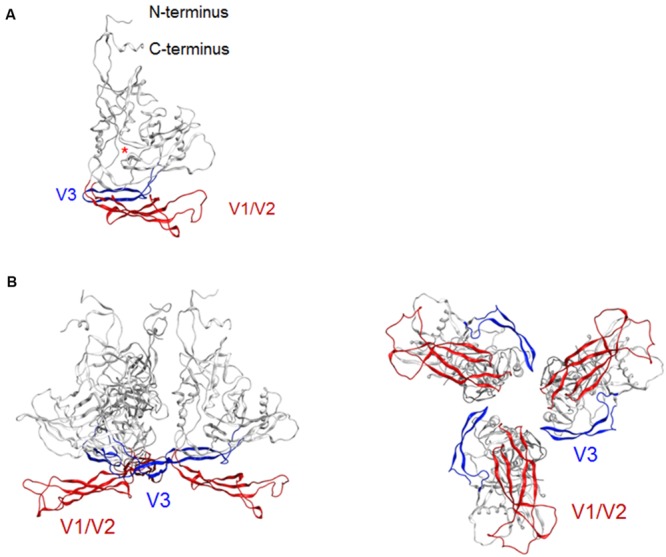
Model for a full-length HIV-1JRFL gp120 in a CD4-free state. (A) Full-length gp120 monomer model. The structure at 50 ns of MD simulation in Figure 1D is shown. Glycans are removed for clear view of the outer domain. A red asterisk indicates the location of CD4-binding loop. (B) Full-length gp120 trimer model. The model was constructed by superposing CD4-free monomer model on the x-ray crystal structure of Env gp140 protein (PDB code: 4TVP; Pancera et al., 2014). Side and top views are shown on the left and on the right, respectively. V1/V2 and V3 regions are highlighted by red and blue colors, respectively.
