Figure 2.
TANGO2-Specific CNV Detection from the Clinical Exome Data and the Spectrum of Mutations in Nine Unrelated Families
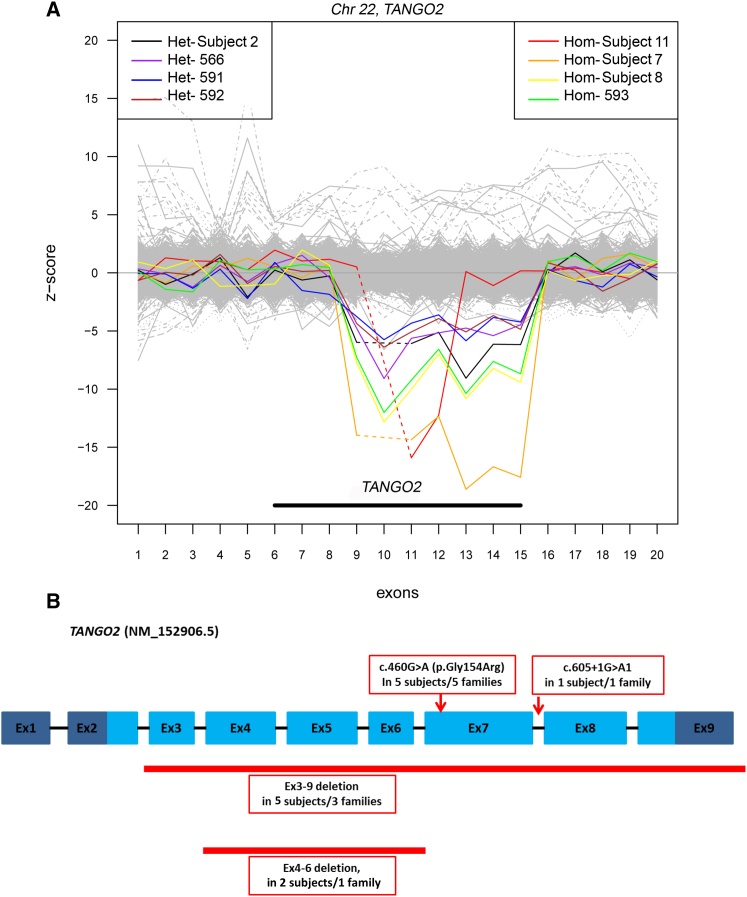
(A) Target Z-score of PCA-normalized read depths for exon targets of TANGO2. Samples with heterozygous or homozygous deletions are shown in various colors on the left and right, respectively. All samples with deletions are depicted with read depths (RD) well below the other samples (gray) (n = 4,334 total samples). Samples with homozygous deletions have RD lower than heterozygous deletions and contain exons with reads per kilobase of transcript per million mapped reads (RPKM) values equal or near 0. Individuals with exons 3–9 homozygous deletions (∼34 kb) and heterozygous deletions are shown in this figure, as subjects 2, 7, and 8. Subject 11 with the smaller exons 4–6 homozygous deletion is represented in red.
(B) TANGO2 transcript (GenBank: NM_152906.5) with mutations and deletions found in our study cohort. Light blue regions represent the coding exons, dark blue regions represent 5′ and 3′ untranslated regions, and black bars represent introns (not to scale). The locations of c.460G>A (p.Gly154Arg) and c.605+1G>A variants are shown. The ∼34 kb deletion that encompasses exons 3 through 9 and the ∼9 kb deletion that includes exons 4 through 6 are shown.