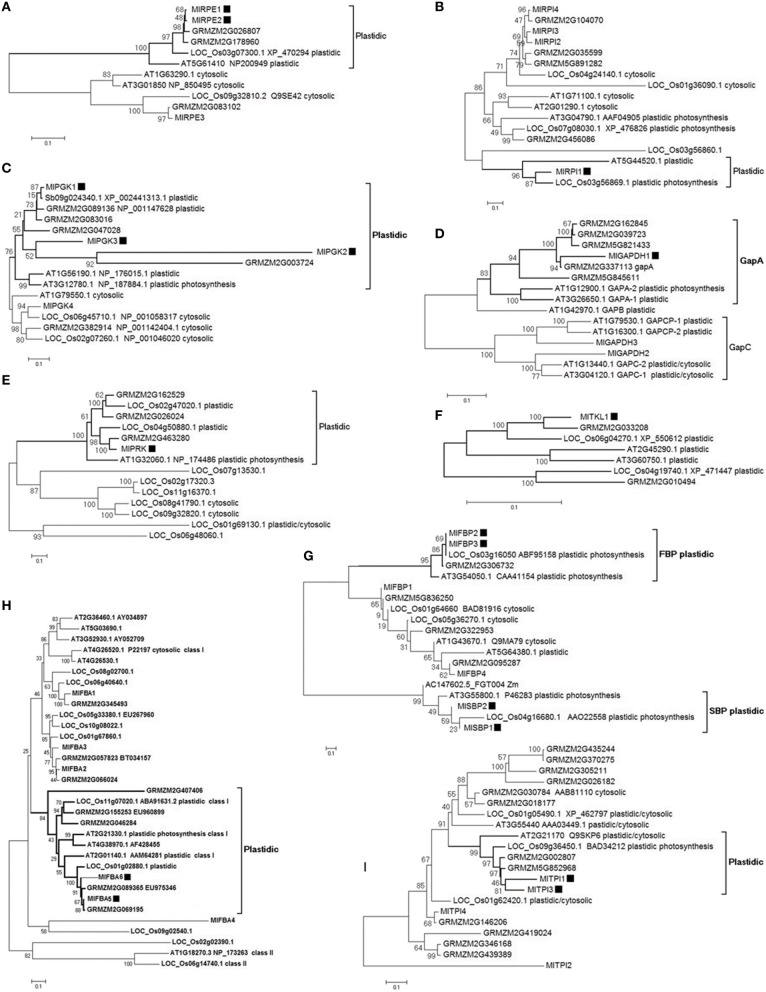
Figure 2.
Neighbor-joining trees of Calvin enzyme transcripts and their isoforms in M. lutarioriparius, S. bicolor, Z. mays, O. sativa, and A. thaliana. Bootstrap percentage values in 500 times are shown as integers. Black square shows the identified M. lutarioriparius transcript involved in Calvin cycle. In the transcript IDs, Ml indicates M. lutarioriparius, Sb indicates S. bicolor, Os indicates O. sativa, GRMZM indicates Z. mays, and AT indicates A. thaliana. (A) RPE, D-ribulose-5-phosphate 3-epimerase; (B) RPI, ribose-5-phosphate isomerase; (C) PGK, phosphoglycerate kinase; (D) GAPDH, glyceraldehyde-3- phosphate dehydrogenase; (E) PRK, phosphoribulokinase; (F) TKL, transketolase; (G) FBP and SBP, fructose bisphosphatase and Sedoheptulose-bisphosphatase; (H) FBA, fructose- bisphosphate aldolase; (I) TPI, triose phosphate isomerase.