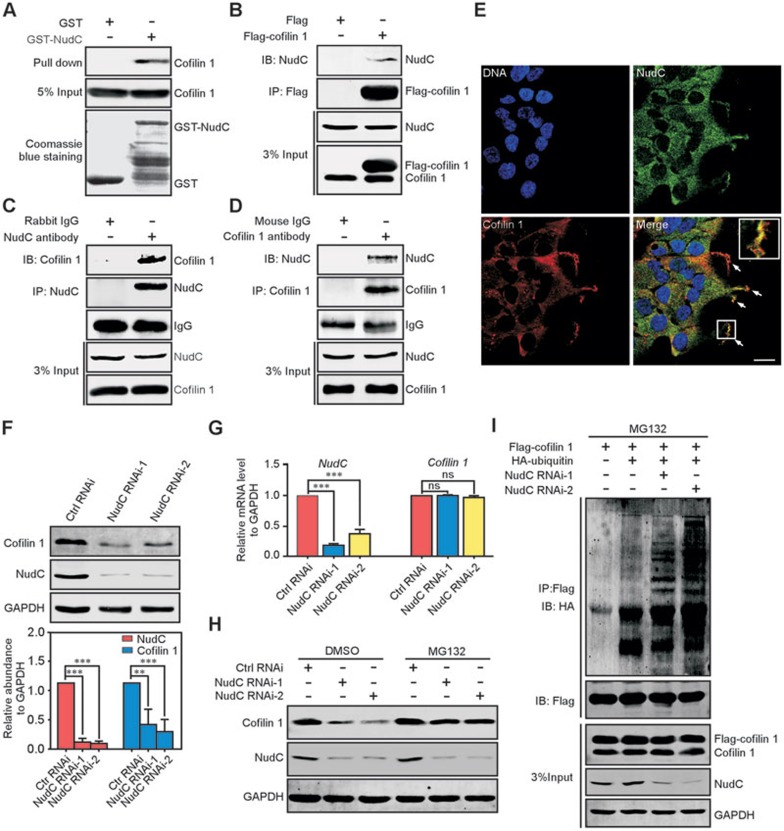
Figure 4.
NudC binds to and stabilizes cofilin 1. (A) GST pull-down assay revealing the interaction between NudC and cofilin 1 in vitro. Input proteins are stained with Coomassie brilliant blue. 5% of total input from RPE-1 cell lysates is shown. (B-D) Co-immunoprecipitation confirms that NudC is associated with cofilin 1 in vivo. RPE-1 cells transfected with the indicated vectors (B) or not (C, D) are subjected to immunoprecipitation with indicated antibodies and western blotting. 3% of total input is shown. (E) Immunofluorescence revealing colocalization of NudC and cofilin 1. DNA is stained with DAPI (blue). Higher magnification of the boxed region is shown in inset. Scale bar, 20 μm. (F) Western analysis showing significantly decreased levels of cofilin 1 in NudC-depleted cells. Band intensities are quantified using Image J software. (G) Quantitative RT-PCR revealing that NudC RNAi has no significant effect on cofilin 1 mRNA. GAPDH is the internal control. (H) Proteasome inhibitor MG132 reverses cofilin 1 degradation induced by NudC depletion. RPE-1 cells treated with or without NudC RNAi were incubated with either 5 μM MG132 or DMSO (vehicle), and subjected to western blotting. (I) NudC depletion induces the ubiquitination of cofilin 1. RPE-1 cells depleted of NudC or not were transfected with the indicated vectors and treated with 5 μM MG132. Immunoprecipitates with anti-Flag antibody from cell lysates are analyzed by immunoblotting. 3% of total input is shown Quantitative data from three independent experiments are shown as mean ± SD. n, sample size. **P < 0.01; ***P < 0.001; ns, not significant (P > 0.05).