Abstract
MicroRNAs (miRNAs) are critical regulators of gene expression, and exert extensive impacts on development, physiology, and disease of eukaryotes. A high degree of parallelism is found in the molecular basis of miRNA biogenesis and action in plants and animals. Recent studies interestingly suggest a potential cross-kingdom action of plant-derived miRNAs, through dietary intake, in regulating mammalian gene expression. Although the source and scope of plant miRNAs detected in mammalian specimens remain controversial, these initial studies inspired us to determine whether plant miRNAs can be detected in Western human sera and whether these plant miRNAs are able to influence gene expression and cellular processes related to human diseases such as cancer. Here we found that Western donor sera contained the plant miRNA miR159, whose abundance in the serum was inversely correlated with breast cancer incidence and progression in patients. In human sera, miR159 was predominantly detected in the extracellular vesicles, and was resistant to sodium periodate oxidation suggesting the plant-originated 2'-O-methylation on the 3′ terminal ribose. In breast cancer cells but not non-cancerous mammary epithelial cells, a synthetic mimic of miR159 was capable of inhibiting proliferation by targeting TCF7 that encodes a Wnt signaling transcription factor, leading to a decrease in MYC protein levels. Oral administration of miR159 mimic significantly suppressed the growth of xenograft breast tumors in mice. These results demonstrate for the first time that a plant miRNA can inhibit cancer growth in mammals.
Keywords: breast cancer, cross-kingdom regulation, miR159, TCF7
Introduction
MicroRNAs (miRNAs), a major family of small RNAs, are ∼23 nt-long single strands of RNA that bind to mRNA transcripts to inhibit their translation1,2. Over the past decade miRNAs have garnered extensive attention due to their unique ability to regulate multiple targets and influence a variety of biological processes including cell growth, metabolism, development, and survival. They play an important role in human diseases, and notably dysregulation of miRNAs has been found in many cancers including breast cancer (BC)3,4. In addition to their intracellular presence, miRNAs are also detected in nearly all biofluids, including serum and plasma, and circulating miRNAs have emerged as potential biomarkers for cancer diagnosis and prognosis5,6,7,8. miRNAs can be secreted through extracellular vesicles (EVs), such as exosomes, shedding vesicles, and apoptotic bodies, or in complexes with protein or lipid-based carriers9,10. Accumulating evidence demonstrates that miRNAs can be transferred via EVs to neighboring or distant cells to modulate cell function11,12,13,14,15,16,17,18,19. Extracellular miRNAs are therefore emerging as a new group of messengers and effectors in intercellular communication.
The biogenesis and acting mechanism of miRNAs display a high degree of similarity between animals and plants, along with evolutionary divergence in the composition and function of miRNA processing machinery2,20. A recent study by Zhang et al. reported that plant-derived miRNAs can be found in human serum. The group demonstrates that the plant miRNA miR168 may be taken up through dietary intake to inhibit the expression of its target low-density lipoprotein receptor 1 in the liver21, providing the first evidence that miRNA in food may influence gene expression in mammalian organs. It is proposed that ingested plant miRNAs are taken up by the gastrointestinal (GI) epithelial cells, which subsequently package the miRNAs into EVs and release them into circulation. A more recent finding by the same group shows that a plant miRNA from honeysuckle is able to inhibit Influenza A replication22, indicating that plant miRNAs may be useful for treating human diseases. Furthermore, another group recently reported that oral administration of three mammalian miRNAs that have been 2'-O-methylated, like plant miRNAs23, resulted in the uptake of theses miRNAs in the intestine and the subsequent reduced intestinal tumor burden24. However, it remains unclear whether ingested miRNAs may exert a therapeutic effect on cancers outside of the GI tract.
On the other side of the controversy, the potential scope of the plant miRNAs detected in mammalian specimens and the real source of foreign miRNA taken up from dietary consumption have been questioned25,26,27,28,29,30,31. Although miRNAs from many exogenous organisms have been reported in human serum32 and breast milk33, several labs were unable to detect their uptake after consumption of various types of food27,28. However, it has been argued that the inability to detect the uptake of plant miRNAs was due to only looking at a few selected miRNAs rather than using sequencing to screen for all plant miRNAs. Furthermore, the cross-kingdom presence and potential function of plant miRNAs in mammals are supported by independent studies from only two different labs in the past three years, highlighting the need for further verification22,34,35,36.
In this study we sought to clarify whether plant miRNAs can be detected in human sera and assess whether they can influence cancer outside of the GI tract such as BC. We found that plant miR159 could be detected in human sera and its levels were inversely correlated with BC incidence and progression. We further identified TCF7 as a mammalian target for miR159 and showed the anti-proliferative function of miR159 in BC cells using in vitro and in vivo models, demonstrating for the first time that a plant miRNA is able to influence BC cell growth.
Results
Plant miR159 can be detected in human sera and tumor tissues, and is associated with breast cancer progression
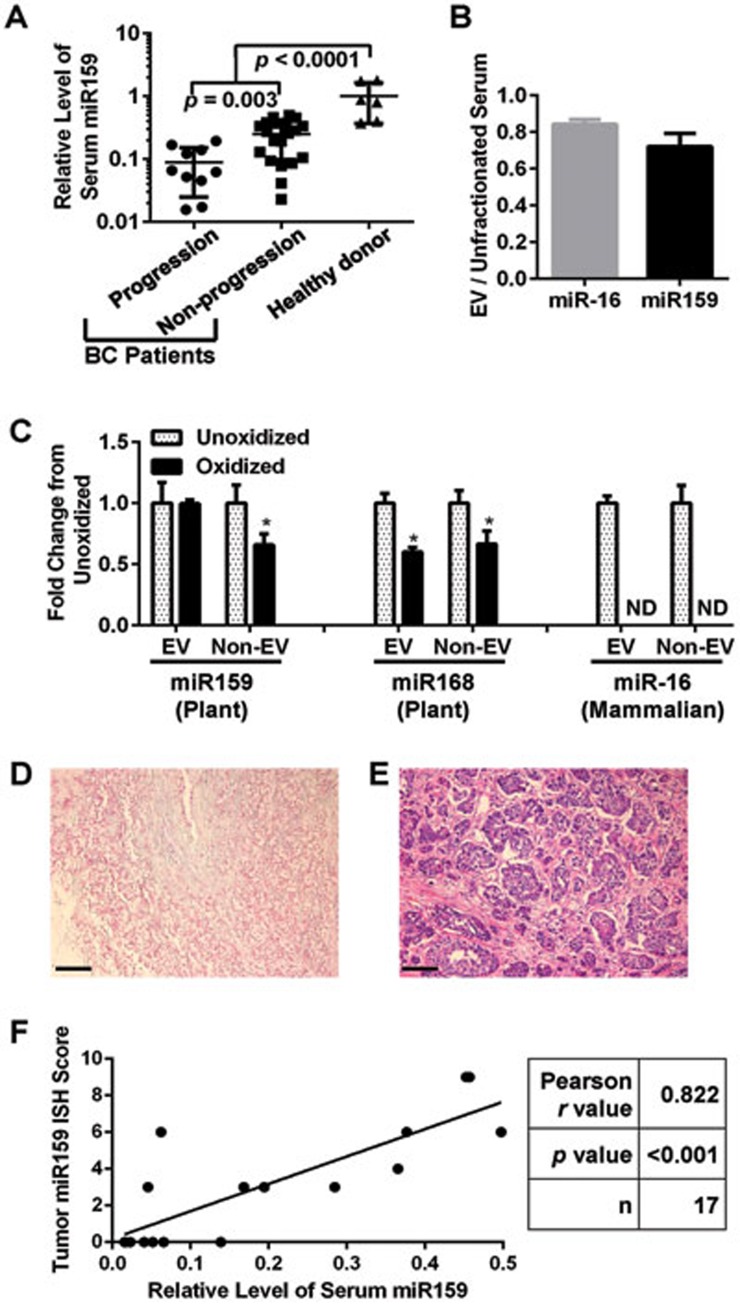
We have previously profiled small RNAs in the sera of 42 stage II-III BC patients by Solexa deep sequencing7. By systemically aligning the detected sequences to the miRNAome of Arabidopsis thaliana and soybean (Glycine max), multiple plant miRNAs were identified but only with sparse counts (Supplementary information, Table S1), possibly due to the 2'-O-methylation on the 3'-terminal ribose in plant-derived miRNAs that causes reduced ligation efficiency of adaptor oligonucleotides37. Among the detected plant miRNAs, ath-miR159a (from Arabidopsis thaliana), gma-miR159a-3p and gma-miR159e-3p (from Glycine max) sharing the identical sequence “uuuggauugaagggagcucua” had the highest counts and were over 6-fold more abundant than other plant miRNAs; therefore we chose to focus on this plant miRNA to see whether it could influence BC progression. Among the Solexa-identified small RNA sequences in patient sera that were aligned to plant miR159, the majority exactly matched to the reference sequence in plants (Supplementary information, Table S2). We further confirmed the presence of miR159 by quantitative PCR (qPCR) using part of these serum RNA samples from BC patients (30 cases available after Solexa) as well as those from individual female healthy donors. Interestingly, circulating miR159 was significantly more abundant in healthy donors in comparison to BC patients (Figure 1A). Furthermore, circulating miR159 was significantly less abundant in BC patients who relapsed with metastatic disease within 3.5 years after serum collection compared to patients who did not have disease progression during 2-6 years of follow-up. In addition, significantly lower levels of circulating miR159 were associated with a negative status of estrogen receptor (ER) or progesterone receptor (PR) expression in the tumor, whereas the expression of HER2 and inflammation status in the tumor were not associated with miR159 levels (Supplementary information, Tables S3 and S4). These results indicate that plant miR159 sequences are present in human serum and are inversely correlated with BC incidence and progression.
Figure 1.
miR159 is detectable in human serum and breast tumor tissue. (A) qPCR for miR159 in the serum of healthy human donors (n = 6) and breast cancer patients who did (n = 10) or did not (n = 20) progress to a higher stage of disease after chemotherapy. Data were first normalized to the level of human miR-16 and then plotted relative to the average level of miR159 in healthy human donors. (B) Enrichment of miR-16 and miR159 in EVs isolated from healthy human donor serum by ultracentrifugation (n = 3). (C) qPCR for the indicated miRNAs with (oxidized) or without (unoxidized) treatment with sodium periodate. RNAs were extracted from EVs isolated from human sera or from human sera depleted of EVs by ultracentrifugation. Data were normalized to miRNA levels of unoxidized samples (n = 3). (D, E) Representative ISH images of two human breast tumors (D) without or (E) with detectable miR159. (F) Scatter plot of ISH scores for miR159 expression in the tumor and the relative serum miR159 abundance shown in A. Scale bar, 100 μm. *P < 0.05. ND, not detectable.
Since miRNAs are primarily transported to distant organs through EV-mediated trafficking14, we assessed whether miR159 is located in EVs in human serum. The majority of miR159 found in human serum was detected in an EV-enriched serum fraction obtained by ultracentrifugation (Figure 1B). Similarly, miR-16, a mammalian miRNA used as a control, was present primarily in the EVs. Plant miRNAs bear a single 2'-O-methylation on their 3′ end23, which is necessary for their stability38. Since mammalian miRNAs lack this modification, we used sodium periodate oxidation to confirm that the miR159 in human serum is of plant origin. The 2′-O-methylation makes plant miRNAs resistant to oxidation by sodium periodate; in contrast mammalian miRNAs lack this protection and are oxidized, preventing them from being amplified by the miScript reverse transcription system21. Subsequent qPCR showed that the majority of the plant miRNA miR168, a previously reported plant miRNA in human serum21, and miR159 in human serum were resistant to sodium periodate and thus are likely of plant origin (Figure 1C). In contrast, the mammalian miR-16 was sensitive to sodium periodate and was not detectable after oxidization. Using in situ hybridization (ISH), we further confirmed that miR159 could be detected in human breast tumors and that the miR159 abundance in the tumor was positively correlated to the miR159 abundance in the serum (Figure 1D-1F). These data suggest that the miR159 in human serum is capable of reaching breast tissue.
Since miR159 is a plant miRNA, the most likely source of miR159 in human serum is from dietary consumption. We profiled the abundance of miR159 in several commonly consumed plants and found that broccoli was particularly rich in miR159 (Supplementary information, Figure S1A). Furthermore, the majority of miR159 was still present after cooking, suggesting that it could be taken up from both raw and cooked foods (Supplementary information, Figure S1B).
Synthetic miR159 suppresses proliferation of BC cells
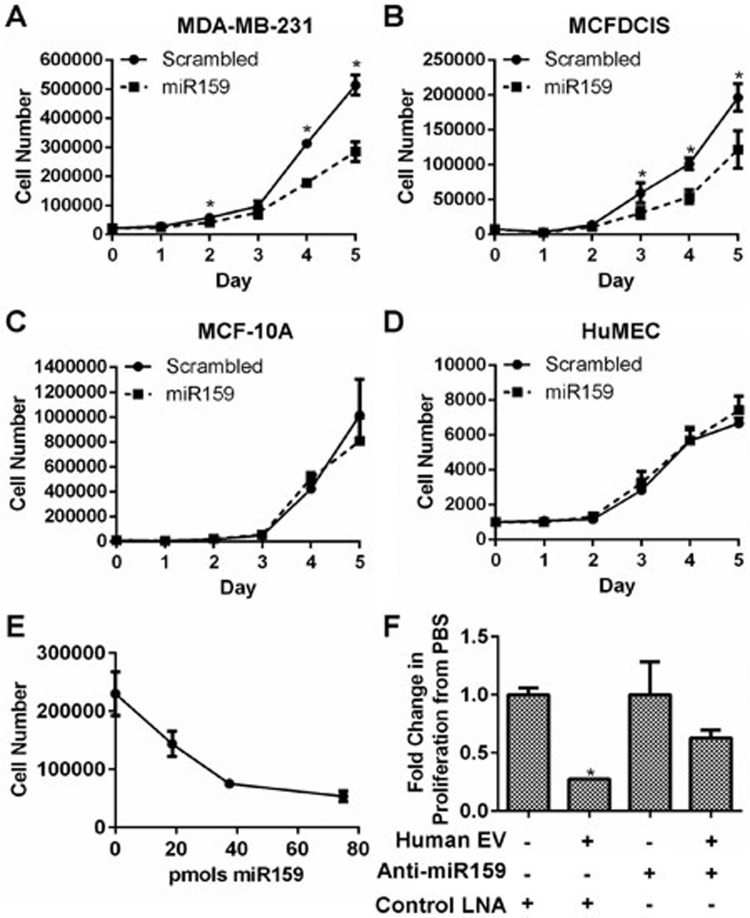
Since we were able to detect miR159 in human sera and breast tumors, we next tested whether miR159 may influence BC cell growth. Cell counting assays demonstrated that a synthetic, double-stranded and 2'-O-methylated miR159 that mimics the duplex of gma-miR159a-3p and gma-miR159a-5p significantly reduced proliferation of the BC cell lines MDA-MB-231 and MCFDCIS, but not the non-cancerous breast cell line MCF-10A or human mammary epithelial cells (HuMECs) (Figure 2A-2D). This growth inhibitory effect of miR159 was dose-dependent in MDA-MB-231 cells (Figure 2E). Since miR159 can be detected in human serum, we tested whether the miR159 in serum is sufficient to inhibit cell proliferation. MDA-MB-231 cells were transfected with either anti-miR159 or control locked nucleic acids (LNAs) and then treated with EVs isolated from healthy human donor serum. Human serum EVs reduced the proliferation of MDA-MB-231 cells, which was partially restored by anti-miR159 LNAs (Figure 2F). Therefore, the miR159 in human serum is capable of inhibiting BC cell proliferation.
Figure 2.
miR159 decreases BC cell proliferation. (A-D) Cell proliferation of breast cell lines transfected with a miR159 mimic or scrambled control oligo. Cell proliferation was determined by cell counting (n = 3). (E) Cell counting of MDA-MB-231 cells transfected with the indicated dose of miR159. Cell were counted 4 days post-seeding (n = 3). (F) Cell counting of MDA-MB-231 cells transfected with anti-miR159 LNAs or control LNAs and treated with human serum EVs. Cells were treated with EVs isolated from 4 ml of pooled human sera or PBS twice over 4 days (n = 3). *P < 0.05.
miR159 binds to and targets transcripts of human TCF7
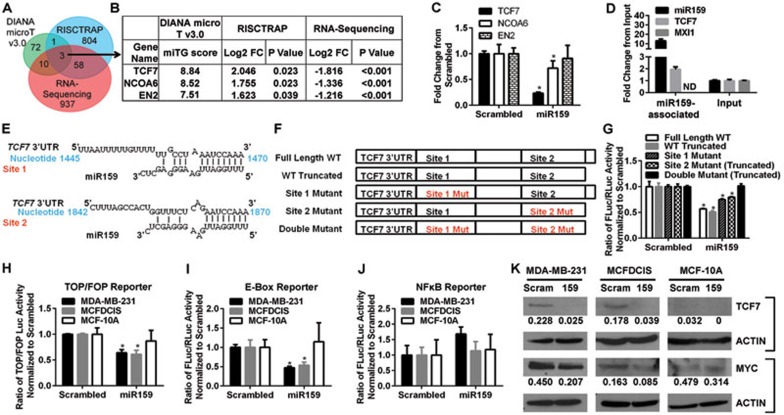
In plants, the miR159 family controls plant growth, development, and morphology by targeting transcription factors MYB33 and MYB6539; however, neither miR159 nor its plant targets have any significant mammalian homolog. To identify what transcripts miR159 is targeting in humans, we used three independent methods, including computational prediction using software DIANA microT v3.0, RNA-Seq-based gene expression profiling of MDA-MB-231 cells transfected with synthetic miR159, and the RNA-induced silencing complex (RISC)-trap (RISCTRAP) assay40 followed by sequencing, to identify endogenous transcripts associated with miR159 (Figure 3A, 3B and Supplementary information, Table S5). Three genes, transcription factor 7 (TCF7), nuclear receptor coactivator 6 (NCOA6), and engrailed-2 (EN2), were identified as putative miR159 targets by all three methods. TCF7 is a TCF/LEF family transcription factor of the Wnt signaling pathway that is upregulated in BC41,42, is a target of several tumor suppressor miRNAs43, and can drive BC cell proliferation44,45,46. NCOA6 is a transcriptional coactivator that is upregulated in BC47,48 and may influence a variety of processes including cell growth, development, and wound healing49. EN2 is a homeobox transcription factor that is upregulated in a subset of BCs and can contribute to transformation if ectopically overexpressed50. Among these three putative miR159 targets, we focused on TCF7, as it has the highest miTG score calculated by the DIANA microT v3.0 prediction software, the highest fold enrichment in the RISCTRAP pull-down assay using biotinylated miR159, and the strongest fold decrease in RNA-Seq and qPCR analyses in MDA-MB-231 cells transiently transfected with miR159 (Figure 3B-3C). Using qPCR, we further confirmed that the transcripts of TCF7 were associated with miR159 in the RISCTRAP assay, whereas those of MXI1, a control gene, were not (Figure 3D).
Figure 3.
miR159 directly targets human TCF7. (A) Overlap of putative miR159 targets in humans identified by RNA-Seq of MDA-MB-231 cells transfected with miR159 mimic, Diana microT v3.0 target prediction software, and the RISCTRAP miR159 association assay. (B) Fold change and statistical significance of the three target genes identified by all three methods. (C-D) qPCR validations of the regulation of the mRNA levels of putative miR159 targets by miR159 in MDA-MB-231 cells (C) and the association of miR159 with TCF7 mRNA in RISCTRAP assay (D). (E) The two putative miR159-binding sites in TCF7 3′UTR identified by Diana microT v3.0. (F) Schematic of the TCF7 miR159-binding site reporter constructs. (G) Dual-luciferase assay in MD-MB-231 cells co-transfected with miR159 or scrambled control oligos and the reporter constructs from F. (H-J) Dual-luciferase assays in the indicated breast cells transfected with miR159 or scrambled control oligos for TOP/FOP (H), E-box (I) or NFκB reporter (J) activity. In C, D, G, and H-J, data shown represent mean ± SD (n = 3). (K) Western blot assays to evaluate TCF7 or MYC expression in breast cell lines transfected with miR159 or scrambled control oligos. Band intensities were measured using ImageJ and were normalized to ACTIN. *P < 0.05. ND, not detectable.
DIANA microT v3.0 identified 2 putative miR159-binding sites in the TCF7 3′UTR (Figure 3E). To verify whether these sites indeed mediate miR159's regulatory effect on TCF7 expression, the TCF7 3′UTR carrying wild-type (WT) or mutant miR159-binding sites was cloned into a luciferase reporter plasmid (Figure 3F). MDA-MB-231 cells were transfected with the reporter plasmids along with synthetic miR159 or a scrambled control to verify the effect of each binding site (Figure 3G). Both full-length and truncated WT 3′UTR plasmids were generated as the mutagenic cloning of site 2 resulted in a 160-bp 3′-end truncation. miR159 was able to reduce the luciferase activity of both WT reporters to a similar degree, indicating that the 160 base pairs lost in the WT truncated 3′UTR are not necessary for miR159 binding. Mutating either of the binding sites resulted in partial restoration of luciferase activity, and complete restoration occurred when both sites were mutated (Figure 3G). Therefore, miR159 is able to bind to both sites to inhibit TCF7 expression. We further examined the effect of miR159 on the transcriptional activity of TCF and NFκB (control) in BC and normal breast cell lines using the corresponding luciferase reporter assays (Figure 3H and 3J). miR159 was able to reduce the transcriptional activity of TCF in MDA-MB-231 and MCFDCIS BC cell lines, but not in the non-cancerous MCF-10A cells (Figure 3H). In contrast, miR159 was not able to alter the NFκB activity in any of the cell lines (Figure 3J), demonstrating the specificity of this effect. One of the downstream targets of TCF7 is the E-box-binding protein and potent oncogene MYC. We used the E-box luciferase reporter assay to assess whether miR159 is able to inhibit MYC activity. Similar to the results with the TCF reporter, miR159 was able to reduce the activity of MYC in MDA-MB-231 and MCFDCIS BC cell lines, but not in the non-cancerous MCF-10A cells (Figure 3I). Western blotting confirmed that miR159 was able to reduce TCF7 and MYC protein levels in BC cells but not in MCF-10A cells (Figure 3K). The lack of a miR159 effect in the non-cancerous MCF-10A and HuMEC cells is likely due to their very low or undetectable level of TCF7 (Figure 3K and data not shown).
miR159 inhibits BC cell growth by targeting TCF7
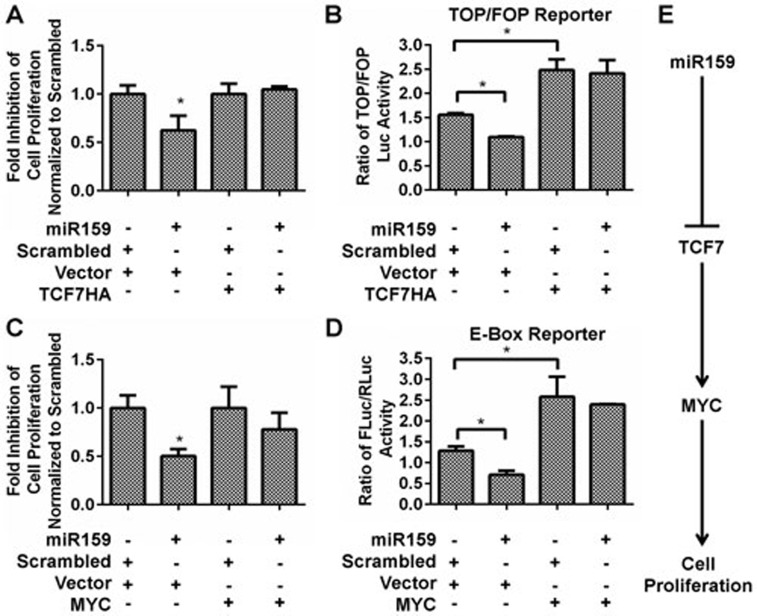
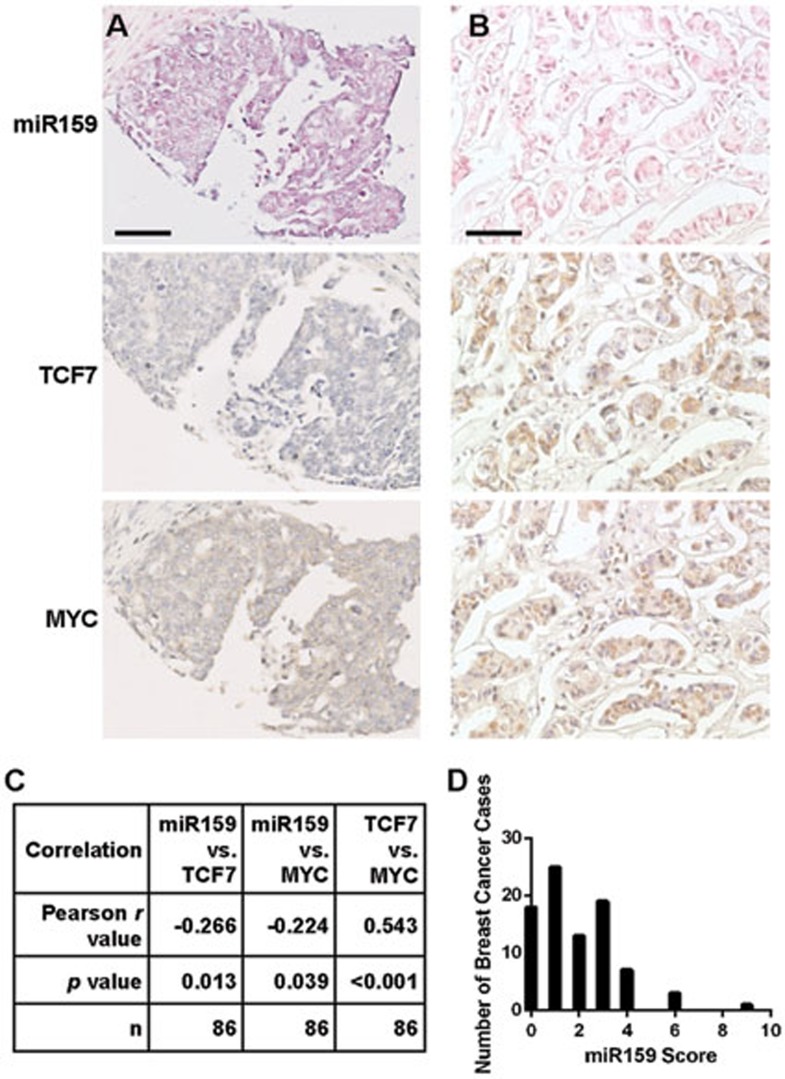
To determine whether miR159 inhibits BC cell growth through targeting TCF7, MDA-MB-231 cells were co-transfected with a pcDNA3-HA-TCF1 plasmid encoding TCF7 without the 3′UTR (TCF7HA) and synthetic miR159. miR159 was able to inhibit proliferation and TCF activity in vector-transfected cells, but failed to do so in cells expressing miR159-resistant TCF7HA (Figure 4A and 4B). Furthermore, miR159 failed to inhibit proliferation or MYC activity in cells overexpressing exogenous MYC (Figure 4C and 4D). These data suggest that miR159 inhibits BC cell proliferation by targeting TCF7, which results in decreased MYC protein level (Figure 4E). We did not observe an increase in proliferation upon forced overexpression of exogenous MYC, possibly due to the saturation of cofactors that inhibit the pro-apoptotic functions of MYC51,52,53,54. In a breast tumor array, we detected a significant inverse correlation between the intratumoral levels of miR159 and either TCF7 or MYC, suggesting that miR159 may target TCF7 in vivo. A positive correlation between TCF7 and MYC levels was also detected in this array (Figure 5).
Figure 4.
miR159 inhibits cell proliferation through targeting TCF7. (A, C) Cell counting of MDA-MB-231 cells co-transfected with miR159 and TCF7HA (A) or MYC (C). Cells were counted 4 days post-seeding. (B) Dual-luciferase assay for TCF reporter activity. MDA-MB-231 cells were co-transfected with miR159, TCF7HA and either TOPFLASH or FOPFLASH plasmids. (D) Dual-luciferase assay for E-box reporter activity. MDA-MB-231 cells were co-transfected with miR159, MYC and either E-box reporter or vector plasmids. In B and D, the luciferase assay was performed 3 days after transfection. (E) Working Model for miR159 inhibition of breast cancer growth. Data shown represent mean ± SD (n = 3). *P < 0.05.
Figure 5.
miR159 level is inversely correlated with levels of TCF7 and MYC in breast tumors. (A, B) Selected images showing ISH for miR159 and IHC for TCF7 and MYC in two BC samples with (A) or without (B) detectable miR159 from a human breast cancer array. (C) Statistical analysis of correlation between miR159 and TCF7 or MYC levels from the human breast cancer array BC08118. (D) Histogram of miR159 scores from the human breast cancer array. Scale bar, 50 μm.
miR159 suppresses breast tumor growth in vivo
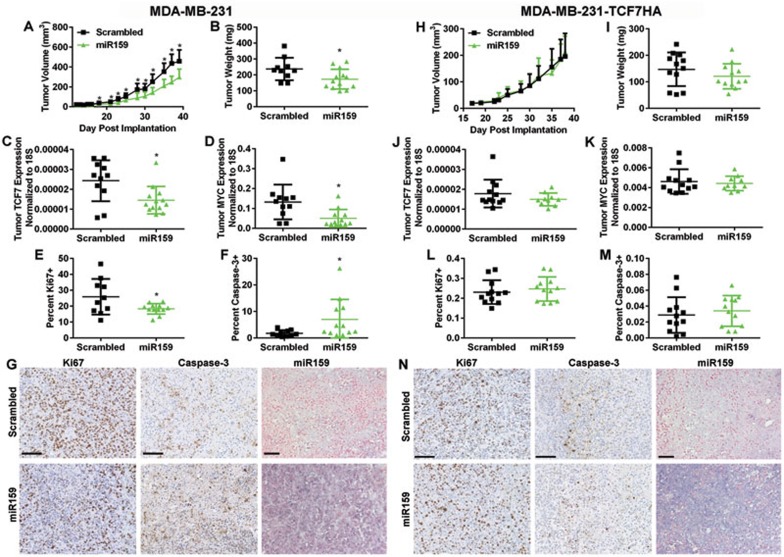
We used a MDA-MB-231 xenograft tumor model to determine whether miR159 inhibits BC growth in vivo. Female NOD-scid IL2Rgnull (NSG) mice were gavage fed daily with synthetic 2'-O-methylated miR159 or scrambled control oligos starting 3 days before injection of MDA-MB-231 cells into the mammary fat pad. The miR159-treated group started to exhibit significantly reduced tumor growth on day 16 and showed significantly decreased tumor weight at the end of the experiment (Figure 6A and 6B). Mice fed with miR159 had significantly reduced TCF7 and MYC expression in their tumors (Figure 6C and 6D), with decreased tumor cell proliferation and increased apoptosis, indicated by Ki67 and cleaved caspase-3 staining, respectively, (Figure 6E-6G). We found that miR159 is indeed detectable, although at much lower levels compared with the GI tract, after a short-term treatment and showed increased accumulation upon a longer treatment in organs outside the GI tract (Supplementary information, Figure S2). Despite this, we were still able to detect miR159 in the xenograft tumors by ISH (Figure 6G). In contrast, miR159 was not able to reduce the growth of MDA-MB-231 tumors stably overexpressing TCF7 without the 3′UTR, and did not significantly alter levels of TCF7 and MYC, or affect proliferation and apoptosis in these tumor cells (Figure 6H-6N). Therefore, the growth inhibitory effects of miR159 are dependent on the TCF7 3′UTR, demonstrating that miR159 targets TCF7 in vivo to reduce breast tumor growth.
Figure 6.
miR159 decreases tumor growth through targeting TCF7 in vivo. (A) MDA-MB-231 tumor growth measured by calipers in mice gavage fed with miR159 or scrambled control oligos. (B) Final tumor weight of MDA-MB-231 tumors. (C, D) qPCR analysis of TCF7 (C) and MYC (D) expression in MDA-MB-231 tumors. (E, F) Quantification of IHC staining signals for Ki67 (E) and cleaved caspase-3 (F) in MDA-MB-231 tumor slides. (G) Representative IHC/ISH images for Ki67, cleaved caspase-3, and miR159 in MDA-MB-231 tumors. In A-G, MDA-MB-231 xenograft mice were fed with miR159 (n = 13) or scrambled control oligos (n = 11). (H) MDA-MB-231-TCF7HA tumor growth measured by calipers in mice gavage fed with miR159 or scrambled control oligos. (I) Final tumor weight of MDA-MB-231-TCF7HA tumors. (J, K) qPCR analysis of TCF7 (J) and MYC (K) expression in MDA-MB-231-TCF7HA tumors. (L, M) Quantification of IHC staining signals for Ki67 (L) and cleaved caspase-3 (M) in MDA-MB-231-TCF7HA tumor slides. (N) Representative IHC/ISH images for Ki67, cleaved caspase-3, and miR159 in MDA-MB-231-TCF7HA tumors. In H-N, MDA-MB-231-TCF7HA xenograft mice were fed with miR159 (n = 12) or scrambled control oligos (n = 12). Scale bar, 100 μm. *P < 0.05.
Discussion
The recent discovery of circulating RNAs originating from exogenous species in humans in their intact and functional forms21,32 led to the paradigm-shifting hypothesis that some RNAs may exert cross-kingdom regulatory effects on physiological processes across different species. If proven true, certain dietary miRNAs from plants and other species may serve as highly affordable and powerful means of treatment with minimal inconvenience to patients. Towards this end, we set out to find plant miRNA sequences existing in human sera that potentially associate with BC, and identified miR159 which (using a synthetic mimic) targets TCF7 to inhibit the proliferation of cells whose growth is dependent on TCF7 such as the BC cells MDA-MB-231. Therefore, miR159 may serve as a potential therapeutic agent to improve current treatment for BCs that require TCF7. It remains to be determined whether plant miRNAs with therapeutic value, such as miR159, can be taken up from certain types of dietary sources at levels that are sufficient to exert a physiological impact on human consumers. Nevertheless, our study using a BC model clearly indicates the anti-tumor effect of orally administered synthetic miR159 in its naturally existing form with the plant-specific 2'-O-methylation, suggesting the feasibility of using synthetic forms of plant miRNAs as dietary supplements in the treatment of human cancers, including those outside of the GI track.
To address whether a physiological dose of miR159 was achieved in our xenograft experiments, we compared the abundance of miR159 in sera of mice receiving equal doses of miR159 as those shown in Figure 6 with sera from 2 lots of pooled human serum samples. The average abundance of miR159 in the xenograft tumor-bearing mice gavage fed with miR159 was 524.3 ± 234.0 fmols/ml serum. Our pooled human serum had 27.5 ± 6.4 fmols/ml of miR159. After normalizing the mouse serum abundance to body surface area to account for the dose differences between mouse and human55, the mouse dose is the equivalent of 42.5 ± 19.0 fmol/ml in humans. Considering that the pooled human serum is expected to be taken from a mix of people who may consume food with variable miR159 abundance and that people who consume food rich in miR159 likely have a higher concentration of miR159 than in our pooled human serum, we believe that the miR159 serum concentration in our xenograft experiments is an acceptable approximation of a typical physiological dose. Our data therefore suggest that dietary plant miR159 may be able to influence BC growth.
The dietary uptake of natural plant miRNAs in mammals has been viewed with skepticism due to the difficulty in observing the uptake of several individual miRNAs from different sources27,28,29,30. This difficulty may be attributed to using PCR to search for selected miRNAs rather than sequencing31 or due to studying a source that does not permit significant uptake of plant miRNAs. Our detection of miR159 among other plant miRNAs in Western human sera from our sequencing data set suggests that plant miRNAs can be taken up through dietary intake. However, the abundance of these miRNAs was very low, with < 40 reads for the most abundant plant miRNA miR159. This could be due to the 2'-O-methylation on the 3′-terminal ribose in plant-derived miRNAs that is known to reduce the ligation efficiency of adaptors used in deep sequencing37, although it remains possible that naturally there is very little stable accumulation of dietary plant miRNAs in the serum. By PCR, we were able to detect miR159 in multiple lots of pooled human sera from healthy donors and in individual sera of BC patients. In an effort to examine the physiological relevance of natural forms of miR159 in human blood, we observed that EVs isolated from healthy donor sera inhibited BC cell growth, which was partially restored by an anti-miR159 LNA, but not a control LNA, indicating that the plant miR159 in normal human serum indeed has anti-tumor activity (Figure 2F).
Several labs have used PCR to note the absence of individual plant miRNAs that were abundant in a given source before ingestion27,28,29; however, our sequencing data as well as others have observed that the plant miRNAs that are taken up after consumption are not necessarily those that are most abundant in the plant (Supplementary information, Table S1)21,22. Therefore, there is likely a selective mechanism for the uptake of specific plant miRNAs in mammals. Furthermore, it should be noted that PCR is dependent on both the concentration of a given gene in the sample as well as the efficiency of the reaction, so caution should be applied when comparing the abundance of two genes with each other. Although our current study along with the previous ones by other groups show that plant miRNAs may regulate human gene expression and even impact human diseases such as cancer, they also intensify the urgency to understand the effects of food-derived miRNAs in human health and disease, for which little evidence exists for one way or another. It is therefore critical to examine the uptake of plant miRNAs from food and the physiological effects of food-derived plant miRNAs in animals using strictly controlled experiments in future studies. It is also important to evaluate the scope of uptake of plant small RNAs (both miRNAs and siRNAs) from food and to understand the molecular basis for the uptake and action of plant-derived small RNAs in animals. These future studies would provide definitive evidence for the highly controversial but potentially revolutionary concept of small-RNA-mediated cross-kingdom action in human-environment interactions, as well as the etiology, prevention, and treatment of many human diseases, such as cancer.
Materials and Methods
Cells, plasmids, and oligos
The human BC cell line MDA-MB-231 and the non-cancerous cell line MCF-10A were obtained from American Type Culture Collection (ATCC; Manassas, VA) and cultured in the recommended media in a humidified 5% CO2 incubator at 37 °C. MCF10DCIS.com (MCFDCIS) cells were purchased from Asterand (Detroit, MI) and cultured in MCF-10A media. HuMECs were purchased from ATCC and cultured in HuMEC Ready Medium (Fisher Scientific; Pittsburgh, PA). The pcDNA3-HA-TCF1 plasmid (TCF7HA) and the TCF luciferase reporter plasmids TOPFLASH (M50 Super 8× TOPFlash) and FOPFLASH (M51 Super 8× FOPFlash (TOPFlash mutant))56, which contains seven TCF/LEF-binding sites inserted into the Mlu1 site of the pTA-Luc vector, were obtained from Addgene (Cambridge, MA). The E-box luciferase reporter plasmid was constructed by inserting four tandem copies of the canonical E-box sequence “CACGTG” upstream of the luciferase gene in pGL3-Promoter vector (Promega; Madison, WI). The NFκB luciferase reporter was generated by cloning the NFκB-binding site into a luciferase reporter construct and was a kind gift from Dr Amy Chow. The pBabe-MYC plasmid was a kind gift from Dr Mei Kong. The CMV-Flag-HA-dnGW182-IRES-puro plasmid was a kind gift from Dr Richard H Goodman. PCR primers used in TCF7 3′UTR cloning are: 5′-ATAATTCCTCGAGAATGCATTCGGTACTTACCCG-3′ (Xho1 forward primer), 5′-TTGCAATGCGGCCGCGGCTTTGAAAAACAAACCCAG-3′ (Not1 reverse primer), 5′-TTTGCCTAAAGAGAAAGAAAAAGG-3′ (site 1 mutant forward primer), 5′-CCTTTTTCTTTCTCTTTAGGCAAA-3′ (site 1 mutant reverse primer), 5′-TTGCAATGCGGCCGCTTTCTCTTCTGAGAAACCAGT-3′ (site 2 mutant Not1 reverse primer), 5′-TTGCAATGCGGCCGCTTTGGATTCTGAGAAACCAGT-3′ (site 2 WT Not1 primer). The TCF7 full-length WT, WT truncated, and site 2 mutant plasmids were generated using the Xho1 forward primer and Not1 reverse primer, site 2 WT Not1 reverse primer, or site 2 mutant Not1 reverse primer, respectively, followed by insertion into the psiCheck-2 plasmid with Xho1 and Not1. The site 1 mutant plasmid was constructed by combining two PCR fragments generated by the Xho1 forward primer and site 1 mutant reverse primer (fragment 1) and the site 1 mutant forward primer and Not1 reverse primer (fragment 2). A site 1 mutant insert was generated using the two PCR fragments mixed at equal molar ratio as the template for a second-round PCR with the Xho1 forward primer and Not1 reverse primer. The double-mutant plasmid was generated using the same strategy as the site 1 mutant with the site 2 mutant plasmid as the template and using the site 2 mutant Not1 reverse primer in place of the Not1 reverse primer. All generated plasmids were confirmed by sequencing. miR159 mimic oligos for in vitro experiments were made by annealing sense and anti-sense strands to create a double-stranded miRNA duplex: miR159 sense 5′-UUUGGAUUGAAGGGAGCUCUmA-3 (“m” denotes 2'-O-methylation), miR159 anti-sense 5′-GAGCUCCUUGAAGUCCAAUUmG, scrambled control sense 5′-GGGACUAGCGGUUAGUGAUAmG-3′, scrambled control anti-sense 5′-CUAUCACUAACCGCUAGUCCmC-3′, miR159-biotin sense 5′-P-UUUGGAUUGAAGGGAGCUCUAUU-biotin-TEG-3′, miR159-biotin anti-sense 5′-P-UAGAGCUCCCUUCAAUCCAUAAG-3′. The miR159-biotin oligo used in the RISCTRAP assay was designed as a double-stranded oligo with a biotin group attached by a TEG linker on the sense strand, which has been reported to not interfere with miRNA function57. Single-stranded oligos were used for in vivo experiments: miR159 5′-UUUGGAUUGAAGGGAGCUCUmA-3′ and scrambled control 5′-UAGUUUGGUAAGGGAGCUCUmA-3′. Anti-miR159 inhibitory LNAs were ordered from Exiqon (Woburn, MA). The negative control A LNA (Exiqon) that does not target any known sequence was used as a control.
Clinical specimens
Human patient serum specimens were obtained from consented patients at City of Hope Medical Center (Duarte, CA) under approved institutional review board protocols. Supplementary information, Table S1 indicating the plant miRNA alignment result is derived from our previously published serum smRNA-seq data set that includes 42 female stage II-III BC patients participating in neoadjuvant chemotherapy clinical trials at City of Hope7. All serum samples were collected at diagnosis before any treatment. Upon diagnosis, all patients received conventional chemotherapy lasting 4-6 months followed by surgical resection of the tumor. Among the 42 patients, 7 received docetaxel/doxorubicin/cyclophosphamide (TAC) treatment regimen (group A) and 12 received doxorubicin/cyclophosphamide (AC) treatment followed by carboplatin and nab-paclitaxel (group B). The other 23 patients had HER2+ BC, and were given the same regimen as for group B but with addition of trastuzumab (group C). One of the HER2+ patients was likely stage IV based on the presence of pleural effusion at diagnosis, which later was documented to be malignant. For PCR analysis of circulating miR159 in Figure 1A, 30 out of the 42 serum RNA samples that were available after smRNA-seq were examined, including 10 cases that later developed systemic recurrence and 20 cases that had not developed systemic relapse. The two groups exhibit balanced age, hormone receptor and HER2 expression, treatment regimen, sample collection time, and follow-up time. Among these 30 cases shown in Figure 1A, 17 had paired primary tumors available for the correlation analyses in Figure 1D-1F. The clinical and histopathological factors of all patients are summarized in Supplementary information, Table S3. Serum RNA was extracted using TRIZOL LS reagent (Life Technologies; Grand Island, NY) and subjected to reverse transcription (RT)-qPCR for miR159 and miR-16 as described7. Individual and pooled healthy female donor human sera were purchased from Innovative Research (Novi, MI). The BC tissue array was purchased from US Biomax (Cat #BC08118; Rockville, MD).
EV isolation
Human serum was spun down at 500× g for 5 min in a tabletop centrifuge at 4 °C to remove any cellular contaminants. The supernatant was spun at 18 700× g for 20 min in a Sorvall Lynx 6000 (Fisher Scientific) centrifuge to remove cellular debris. EVs were pelleted by spinning at 110 000× g for 70 min at 4 °C in a LE-80 ultracentrifuge (Beckman Coulter; Palo Alto, CA). EV-depleted serum was removed and the EV pellet was washed with PBS and spun again at 110 000× g for 70 min at 4 °C. The supernatant was removed by aspiration and the pellet was resuspended in a small volume of PBS.
Sodium periodate qPCR and RISCTRAP assay
RNA from 2 ml of pooled healthy donor serum was extracted using TRIZOL LS. RNA was resuspended in 5 μl RNase-free H2O and incubated with 95 μl 10 mM sodium periodate (Fisher Scientific) for 40 min in a covered ice bucket. RNA was precipitated by adding 1 ml 100% ethanol (Pharmco-AAPER; Brookfield, CT) + 1 μl glycogen (Roche Diagnostics Corporation; Indianapolis, IN) and incubating on ice for 20 min. RNA was pelleted by spinning at 12 000× g for 15 min at 4 °C in a benchtop centrifuge. To remove residual sodium periodate, the resulting pellet was washed with 1 ml 100% ethanol followed by another 12 000× g spin for 15 min at 4 °C. The pellet was washed with 1 ml 75% ethanol and spun down again at 12 000× g for 15 min at 4 °C in a benchtop centrifuge. To remove residual ethanol, the pellet was air dried at room temperature for 5 min. The RNA was resuspended in RNase-free H2O followed by miScript RT-qPCR as stated below. The RISCTRAP assay was performed as previously described40 using biotinylated miR159 and anti-biotin beads (Sigma-Aldrich; St. Louis, MO) to pull down miR159 and associated RNAs, rather than assess total RNA. RNA-Seq was performed to identify miR159-associated RNAs which were compared with input total RNAs.
RNA-Seq and bioinformatics
Solexa sequencing was performed by the City of Hope Integrative Genomics Core using RNA samples from MDA-MB-231 cells transfected with miR159 mimic or scrambled control, and using miR159-associated and input RNAs from the RISCTRAP assay. RNA was reverse-transcribed into cDNA and sonicated. After fragmentation, cDNA fragments with sizes between 130-280 bp were isolated from polyacrylamide-urea gels. The isolated cDNA fragments were ligated with a 3′-linker with a barcode and a 5′-linker. The ligated material was amplified by PCR and then analyzed on a HiSeq2500 (Illumina; San Diego, CA) for parallel sequencing. Sequences were aligned to human genome assembly hg19 and unaligned reads were extracted for plant miRNA profiling. These unaligned reads were aligned to plant miRNAs originated from miRBase v18 using Bowtie not allowing mismatches. The resulted alignment results were then imported into R and expression level of each plant miRNA in each sample was summarized. Putative targets of miR159 were identified using the DianaScan micro T v.3.0 software. Identified putative targets were compared with genes downregulated by miR159 in RNA-Seq data and targets enriched in the RISCTRAP assay.
RNA extraction and RT-qPCR
RNA from cellular samples was extracted using Trizol (Life Technologies) according to manufacturer's instructions. Plant samples were frozen in liquid nitrogen and ground with a mortar and pestle. An equal weight of ground plant material was used for RNA extraction and all extracted RNA was used for cDNA synthesis using the same method as cellular samples.
All RT from cellular samples was done using the miScript II kit (Qiagen; Valencia, CA) according to manufacturer's instructions and qPCR was done using iQ SYBR Green Supermix (VWR; Radnor, PA) with either U6 (for miRNA; Qiagen) or 18S (for mRNA; forward primer: 5′-CTACCACATCCAAGGAAGCA-3′, reverse primer: 5′-TTTTTCGTCACTACCTCCCCG-3′) as internal controls. PCR primers are: miR159 primer 5′-TTTGGATTGAAGGGAGCTC-3′, miR168 primer 5′-TCGCTTGGTGCAGATCGGG-3′, miR-16 (Qiagen, only in sodium periodate treatment), TCF7 forward primer: 5′-CTGGCTTCTACTCCCTGACCT-3, TCF7 reverse primer 5′-ACCAGAACCTAGCATCAAGGA-3′, MYC forward primer 5′-GGCTCCTGGCAAAAGGTC-3′, MYC reverse primer 5′-CTGCGTAGTTGTGCTGATGT-3′. RT for miR-16 and miR159 from serum samples was conducted using the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies) according to manufacturer's instructions, and qPCR was done using TaqMan Universal PCR Master Mix II, No UNG (Life Technologies) with corresponding Taqman probes with the addition of 5% ethylene glycol and 1% DMSO for improved sensitivity. Standard curves were generated from miR159 (5′-UUUGGAUUGAAGGGAGCUCUmA-3′) and miR-16 (5′-TAGCAGCACGTAAATATTGGCG-3′) synthetic oligos.
In situ hybridization (ISH), immunohistochemistry (IHC), and western blot analysis
ISH and IHC were performed as previously described17. The miR159 detection probes were obtained from Exiqon (Woburn, MA). Ki67 antibodies were obtained from Dako (Carpinteria, CA; MIB-1) and cleaved caspase-3 antibodies were obtained from Cell Signaling Technology (5A1E; Beverly, MA). TCF7 (C63D9) and MYC (D84C12) antibodies were obtained from Cell Signaling Technology. Actin antibodies were obtained from Sigma-Aldrich (AC-15).
Transfection, proliferation, and luciferase assays
Transfection was performed according to manufacturer's instructions using RNAiMAX Lipofectamine (Life Technologies) for oligonucleotide transfection, Lipofectamine 2000 (Life Technologies) for plasmid transfection, and DharmaFECT Duo transfection reagent (GE Healthcare; Lafayette, CO) for oligonucleotide + plasmid transfection. For cell proliferation assays, at 24 h post transfection, the cells were seeded at a density of 22 000 cells per 6-cm dish for cell counting. Cells were counted every day for 5 days for cell numbers. In the experiment using various doses of miR159, cell counting was performed 4 days post seeding. For serum EV treatment with anti-miR159 LNAs, MDA-MB-231 cells were transfected with either anti-miR159 or control LNAs and seeded at 22 000 cells per 6-cm dish 24 h after transfection. Twenty-four hours later, cells were treated with human serum EVs isolated from 4 ml of pooled human sera from healthy donors. The cells received a second dose of EVs 48 h after the first and cells were counted 48 h later. For luciferase assays, 3 days after transfection the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee at City of Hope and in compliance with ethical regulations. Female 6-10-week-old NSG mice were used for all animal experiments. For miR159 distribution experiments, mice were gavage fed daily with 25 mg/kg miR159 or scrambled control oligos dissolved in RNase-free H2O. At the indicated time points, the animals were killed and tissues were preserved in RNAlater (Life Technologies) overnight. Tissues were then homogenized using soft tissue homogenizing kit CK14 (VWR) for RNA extraction. For xenograft tumor experiments, mice were gavage fed daily with 25 mg/kg miR159 or scrambled control oligos for 3 days before the implantation of 2 × 105 MDA-MD-231 or MDA-MD-231-TCF7HA cells into the #4 mammary fat pad. Mice continued to receive daily oral gavage of RNA oligos and tumor size was measured by caliper three times a week for 5 weeks, after which mice were killed.
Statistical analyses
All quantitative data are presented as mean ± SD. For all quantitative data, statistical analyses were performed using Student's t-tests. Values of P < 0.05 were considered significant. The correlation analyses were evaluated by Pearson correlation coefficient (r). Values of P < 0.05 were considered significant.
Author Contributions
ARC performed the majority of the experiments. MYF performed ISH and IHC. GS assembled and provided clinical specimens. JW assisted with mouse experiments. PS assisted with design and synthesis of miRNA mimics. XW performed all bioinformatics analyses. ARC and SEW conceived this study and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
This work was supported by the National Institutes of Health (NIH)/National Cancer Institute (NCI) (R01CA166020and R01CA163586 to SEW), California Breast Cancer Research Program (20IB-0118 to SEW), Breast Cancer Research Foundation-AACR (12-60-26-WANG to SEW), and the City of Hope Women's Cancer Program. Research reported in this publication included work performed in Core facilities supported by the NIH/NCI under grant number P30CA33572. We thank Drs Richard H Goodman, Mei Kong, and Amy Chow for providing plasmid constructs and Drs Xuemei Chen, Susan Kane, Ren-Jang Lin, Mark Boldin, and Marcin Kortylewski for valuable comments, as well as the core facilities at City of Hope for highly professional services.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
miR159 abundance in common foods.
Distribution of miR159 after oral gavage.
Solexa sequencing counts of Arabidopsis thaliana and Glycine max (soybean) miRNAs from the sera of 42 BC patients. miR159 counts are in bold.
Solexa-detected miR159 sequences identified in all patient serum samples in Table S1.
Patient characteristics and sample availability.
Relative levels of miR159 normalized to miR-16 in the sera of breast cancer patients grouped by ER, PR, HER2, and inflammatory status.
Putative miR159 targets identified by Diana micro T v3.0 software, RISCTRAP miR159 association assay, and RNA-seq from MDA-MB-231 cells transfected with miR159.
References
- 1Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 2Chen X. Small RNAs in development — insights from plants. Curr Opin Genet Dev 2012; 22:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857–866. [DOI] [PubMed] [Google Scholar]
- 4Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065–7070. [DOI] [PubMed] [Google Scholar]
- 5Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008; 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008; 110:13–21. [DOI] [PubMed] [Google Scholar]
- 7Wu X, Somlo G, Yu Y, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med 2012; 10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes 2009; 2:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther 2012; 136:169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 2012; 32:326–348. [DOI] [PubMed] [Google Scholar]
- 11Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 2011; 108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012; 18:883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10:1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654–659. [DOI] [PubMed] [Google Scholar]
- 15Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol 2012; 23:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 2010; 38:7248–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Fong MY, Zhou W, Liu L, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol 2015; 17:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014; 25:501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015; 17:816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 2009; 25:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 2012; 22:107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Zhou Z, Li X, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res 2015; 25:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Yu B, Yang Z, Li J, et al. Methylation as a crucial step in plant microRNA biogenesis. Science 2005; 307:932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Mlotshwa S, Pruss GJ, MacArthur JL, et al. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res 2015; 25:521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Tosar JP, Rovira C, Naya H, Cayota A. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA 2014; 20:754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Zhang Y, Wiggins BE, Lawrence C, Petrick J, Ivashuta S, Heck G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics 2012; 13:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Snow JW, Hale AE, Isaacs SK, Baggish AL, Chan SY. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol 2013; 10:1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Witwer KW, McAlexander MA, Queen SE, Adams RJ. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA Biol 2013; 10:1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr 2014; 144:1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 2013; 31:965–967. [DOI] [PubMed] [Google Scholar]
- 31Chen X, Zen K, Zhang CY. Reply to Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 2013; 31:967–969. [DOI] [PubMed] [Google Scholar]
- 32Wang K, Li H, Yuan Y, et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One 2012; 7:e51009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Lukasik A, Zielenkiewicz P. In silico identification of plant miRNAs in mammalian breast milk exosomes — a small step forward? PLoS One 2014; 9:e99963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Liang G, Zhu Y, Sun B, et al. Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr 2014; 2:380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Liang H, Zhang S, Fu Z, et al. Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J Nutr Biochem 2015; 26:505–512. [DOI] [PubMed] [Google Scholar]
- 36Yang J, Farmer LM, Agyekum AA, Hirschi KD. Detection of dietary plant-based small RNAs in animals. Cell Res 2015; 25:517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Munafo DB, Robb GB. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 2010; 16:2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 2005; 15:1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Allen RS, Li J, Stahle MI, Dubroue A, Gubler F, Millar AA. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 2007; 104:16371–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Cambronne XA, Shen R, Auer PL, Goodman RH. Capturing microRNA targets using an RNA-induced silencing complex (RISC)-trap approach. Proc Natl Acad Sci USA 2012; 109:20473–20478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Benhaj K, Akcali KC, Ozturk M. Redundant expression of canonical Wnt ligands in human breast cancer cell lines. Oncol Rep 2006; 15:701–707. [PubMed] [Google Scholar]
- 42Johnson JP, Kumar P, Koulnis M, Patel M, Simin K. Crucial and novel cancer drivers in a mouse model of triple-negative breast cancer. Cancer Genomics Proteomics 2014; 11:115–126. [PubMed] [Google Scholar]
- 43Hrdlickova R, Nehyba J, Bargmann W, Bose HR Jr. Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS One 2014; 9:e86990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121:2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Bilir B, Kucuk O, Moreno CS. Wnt signaling blockage inhibits cell proliferation and migration, and induces apoptosis in triple-negative breast cancer cells. J Transl Med 2013; 11:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene 2006; 25:7492–7504. [DOI] [PubMed] [Google Scholar]
- 47Guan XY, Xu J, Anzick SL, Zhang H, Trent JM, Meltzer PS. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11-q13.2 in breast cancer. Cancer Res 1996; 56:3446–3450. [PubMed] [Google Scholar]
- 48Lee SK, Anzick SL, Choi JE, et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem 1999; 274:34283–34293. [DOI] [PubMed] [Google Scholar]
- 49Mahajan MA, Samuels HH. Nuclear receptor coactivator/coregulator NCoA6(NRC) is a pleiotropic coregulator involved in transcription, cell survival, growth and development. Nucl Recept Signal 2008; 6:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Martin NL, Saba-El-Leil MK, Sadekova S, Meloche S, Sauvageau G. EN2 is a candidate oncogene in human breast cancer. Oncogene 2005; 24:6890–6901. [DOI] [PubMed] [Google Scholar]
- 51Evan GI, Wyllie AH, Gilbert CS, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell 1992; 69:119–128. [DOI] [PubMed] [Google Scholar]
- 52Prendergast GC. Mechanisms of apoptosis by c-Myc. Oncogene 1999; 18:2967–2987. [DOI] [PubMed] [Google Scholar]
- 53Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 1999; 13:2678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Reynolds JE, Yang T, Qian L, et al. Mcl-1, a member of the Bcl-2 family, delays apoptosis induced by c-Myc overexpression in Chinese hamster ovary cells. Cancer Res 1994; 54:6348–6352. [PubMed] [Google Scholar]
- 55Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J 2008; 22:659–661. [DOI] [PubMed] [Google Scholar]
- 56Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 2003; 13:680–685. [DOI] [PubMed] [Google Scholar]
- 57Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods 2007; 43:162–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miR159 abundance in common foods.
Distribution of miR159 after oral gavage.
Solexa sequencing counts of Arabidopsis thaliana and Glycine max (soybean) miRNAs from the sera of 42 BC patients. miR159 counts are in bold.
Solexa-detected miR159 sequences identified in all patient serum samples in Table S1.
Patient characteristics and sample availability.
Relative levels of miR159 normalized to miR-16 in the sera of breast cancer patients grouped by ER, PR, HER2, and inflammatory status.
Putative miR159 targets identified by Diana micro T v3.0 software, RISCTRAP miR159 association assay, and RNA-seq from MDA-MB-231 cells transfected with miR159.