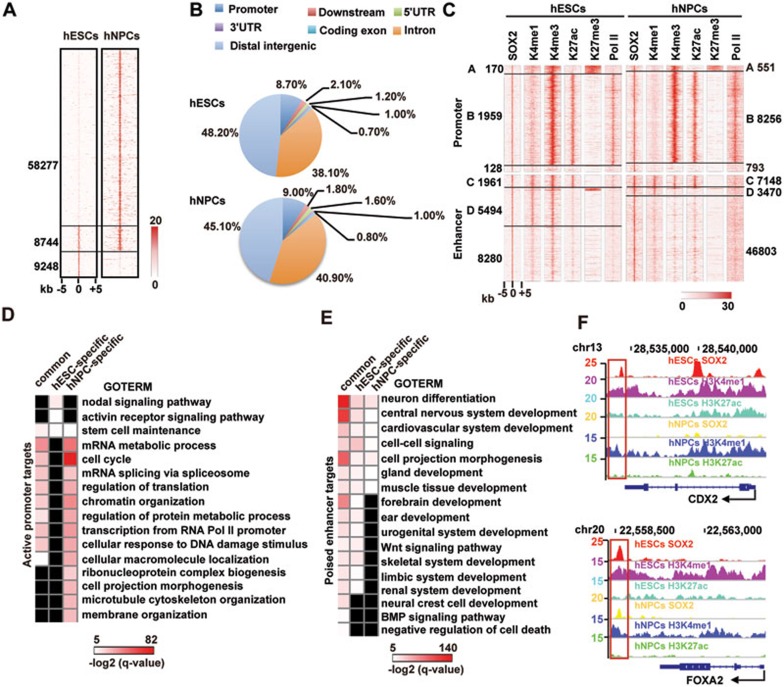
Figure 1.
SOX2-binding sites in hESCs and hNPCs characterized by ChIP-seq data of SOX2, Pol II and major histone modifications. (A) Heatmap of SOX2 and Pol II enrichments in hESCs and hNPCs centered on peaks of enrichment and extended 5 kb in each direction. Numbers of common and specific peaks are indicated. (B) The genomic distribution of SOX2 peaks in hESCs and hNPCs. (C) Heatmaps depicting Pol II, H3K4me3, H3K4me1, H3K27me3 and H3K27ac at regions spanning all SOX2-binding sites (± 5 kb) in hESCs and hNPCs. SOX2 peaks are divided into 4 categories (A, poised promoter by H3K4me3+/H3K27me3+; B, active promoter by H3K4me3+/H3K27me3− C, active enhancer by H3K4me1+/H3K27ac+; D, poised enhancer by H3K4me1+/H3K27ac− and H3K4me1+/H3K27me3+). Numbers of peaks in each category are indicated. (D, E) GO analyses of active promoter-bound genes (D) and poised enhancer-bound genes (E) in hESCs and hNPCs. Benjamini-Hochberg method is applied to adjust the P-values in order to account for multiple testing. Enrichment levels of selected GO terms are marked by −log2(q-value). Missing values are shown as black. (F) Integrative Genomics Viewer screenshots showing the hESC-specific SOX2 peak on CDX2 enhancer and the SOX2 peak on FOXA2 enhancer in both hESCs and hNPCs.