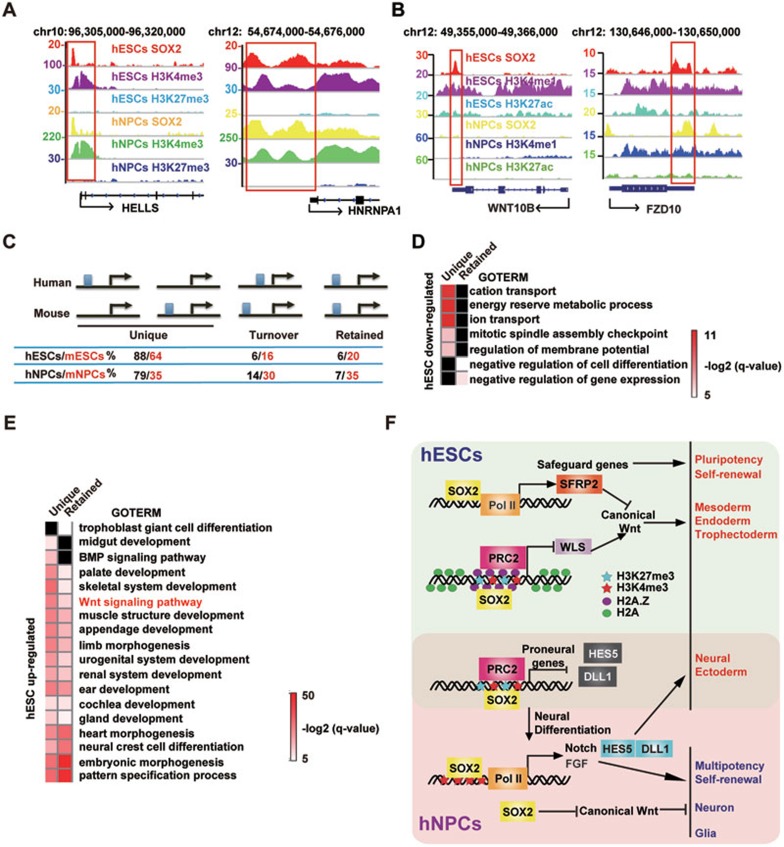
Figure 8.
Comparative analysis between mouse and human SOX2 ChIP-seq reveals conserved and divergent regulation. (A) Integrative Genomics Viewer screenshots of common SOX2 peaks on the active promoters of HELLS and HNRNPA1. (B) Integrative Genomics Viewer screenshots of common SOX2 peaks on the poised enhancer of FZD10 and the hESC-specific peak on the poised enhancer of Wnt10B. (C) A schematic illustration of the classification of retained, turned over and unique SOX2 peaks in ESCs and NPCs of the human and mouse origin. The percentages of corresponding peaks are indicated. (D, E) GO analyses of genes downregulated (D) and upregulated (E) upon SOX2/3 KD in the retained and unique groups. Benjamini-Hochberg method is applied to adjust the P-values in order to account for multiple testing. Enrichment levels of selected GO terms are marked by −log2(q-value). Missing values are marked in black. (F) A working model showing the cell type- and stage-dependent functions of SOX2.