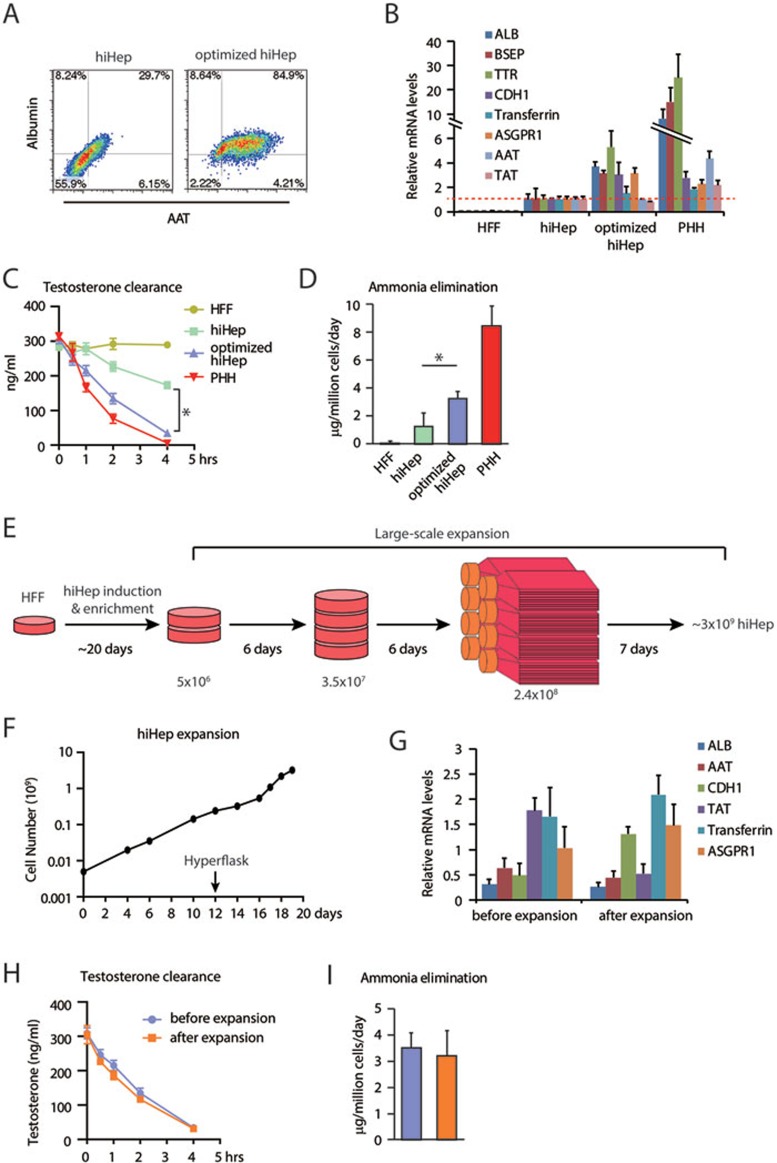
Figure 1.
Characterization of optimized and large-scale expanded hiHeps. (A) Optimized hiHeps show high percentage of albumin and AAT double-positive cells as determined by flow cytometry. (B) q-PCR analyses of hepatic gene expression in optimized hiHeps, including albumin (ALB), ATP-binding cassette subfamily B member 11 (BSEP), transthyretin (TTR), cadherin 1 (CDH1), transferrin, asialoglycoprotein receptor 1 (ASGPR1), AAT, tyrosine aminotransferase (TAT). (C) Testosterone elimination of optimized hiHeps was determined by LC-MS/MS. (D) The ability of eliminating ammonia was measured in optimized hiHeps by enzymatic colorimetric assay. (E) Schematic outline of the large-scale expansion of hiHeps. hiHeps were generated and enriched in 6 cm petri dish and then expanded to 2.4 × 108 in 10 cm dishes. hiHeps were finally expanded in Hyperflasks. (F) Large-scale expansion of hiHep cultures from 5 × 106 to ∼3 × 109 cells in 20 days. (G-I) Large-scale expanded hiHeps maintained hepatic gene expression levels (G), efficient testosterone clearance (H) and ammonia elimination (I). HFF, human fetal fibroblast. PHH, primary human hepatocyte. *P < 0.05, t-test.