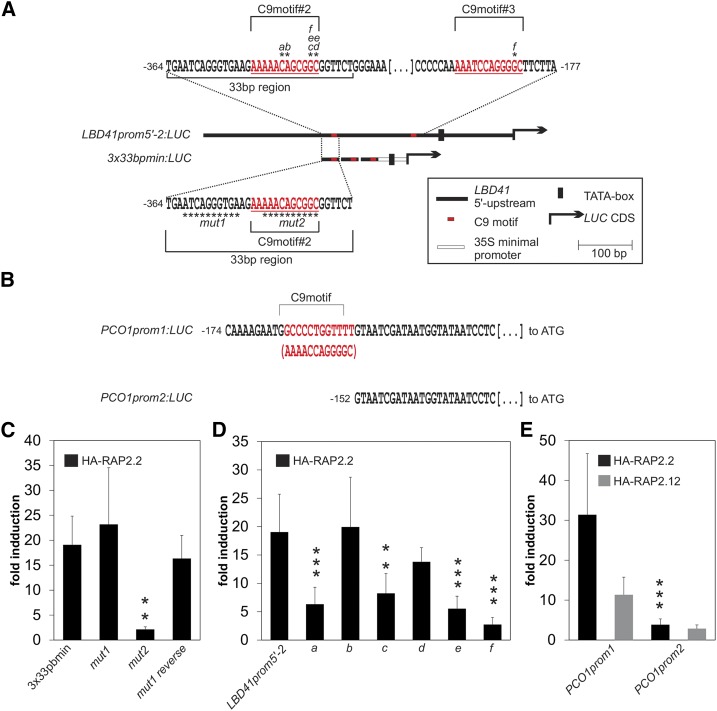
Figure 6.
The C9motif (HRPE) Functions in ERF-VII-Targeted Transactivation.
(A) and (B) Maps of LBD41 and PCO1 promoter fragments cloned 5′ to the LUC coding region with C9motifs and base substitutions or deletions indicated.
(C) Comparison of HA-RAP2.2-induced LUC activity of wild-type and mutated versions of 3x33bpmin:LUC.
(D) Comparison of HA-RAP2.2-induced LUC activity of the 589-bp LBD41prom5′-2:LUC reporter and point mutations in two C9motifs.
(E) Effect of HA-RAP2.2 and HA-RAP2.12 on C9motif-containing and C9motif-less PCO1 promoter versions. All measurements were performed in a transient protoplast transactivation system using p35S:RUC for normalization. Data are means ± sd of six replicates; asterisks indicate significant differences from wild-type-induced promoter activity at ***P < 0.001 and **P < 0.01 (Tukey HSD test). Fold induction, ratio between the transcription factor-induced and basal promoter activity using p35S:HA-GFP. Promoter activity values (LUC activity normalized to p35S:RUC activity) are shown in Supplemental Figure 4.