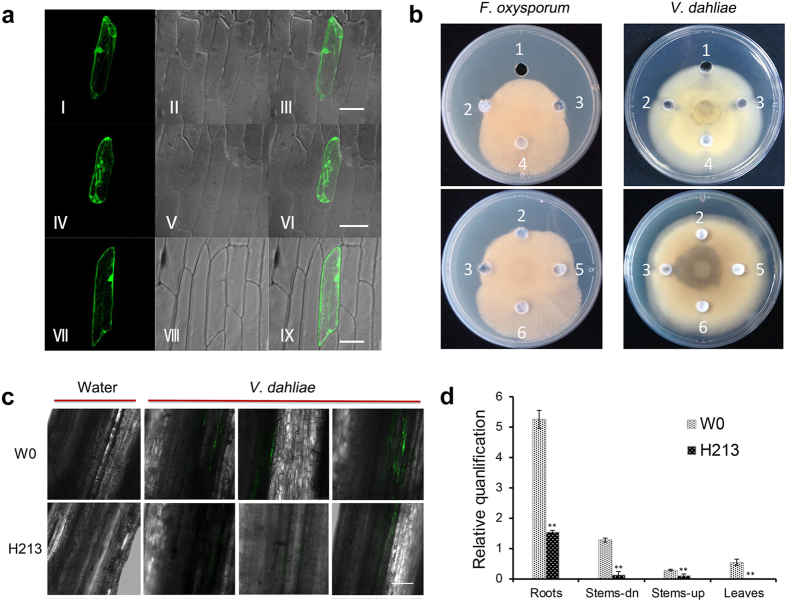
Figure 6. Expression of Hcm1 inhibited the spread of fungal spores in cotton.
(a) Localization of the 35S::Hcm1::GFP fusion in onion epidermal cells. (I–III) Localization of 35S::Hcm1::GFP in onion epidermal cells. (IV–VI) The cell wall and membrane were separated by treatment with 20% sucrose for 15 min. (VII–IX) 35S::GFP in onion epidermal cells (positive control). (I, IV, and VII) Onion cell under excitation at 488nm. (II, V and VIII) Onion cell under bright field. (III, VI and IX) GFP in the onion cell of overlayed images. Scale bars = 50 μm. (b) Antimicrobial activities of CFEPs and transgenic Hcm1 protein against F. oxysporum Fnj1 and V. dahliae V991 on PDA plates. 1–6: 50 μg/ml Carbendazim, 1 mg/ml CFEPs of Hcm1, 1 mg/ml Hcm1-transformed protein, 1 mg/ml CFVPs, 200 μg/ml Hcm1-transformed protein, and 1 mg/ml parent W0 protein. (c) In situ observation of V. dahliae in rhizome connections of transgenic line H213 and parent W0 plants 15 days after inoculation with V. dahliae harboring the GFP gene. At least 15 plants of transgenic lines H213 and parent W0 plants were used in this study. The freehand sections were obtained and checked by laser scanning confocal microscopy. Scale bar = 200 μm. (d) Detection of the V. dahliae biomass in transgenic line H213 and parent W0 plants using qRT-PCR. DNA was extracted from roots, the lower half of stems (stem-dn), the upper half of stems (stem-up) and the first leaves of plants 15 days after inoculation with V. dahliae. The relative average fungal biomass is shown with standard errors. Asterisks indicate significant differences when compared with colonization of the parent W0 plants (**P ≤ 0.01, by student’s t test).