Fig. 3.
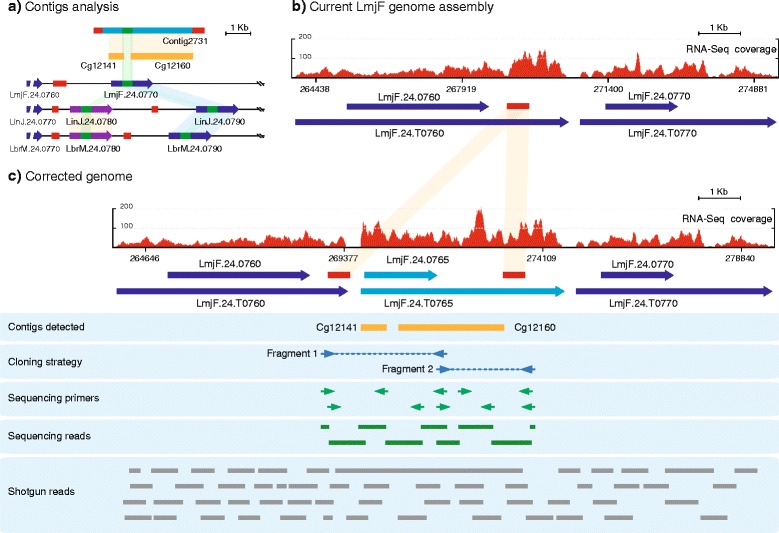
Uncovering of a new gene (LmjF.24.0765) after reconstruction of the intergenic region between genes LmjF.24.0760 and LmjF.24.0770. a Schematic representation of the sequence homology existing between the de novo contigs (Cg12141 and 12160) and Contig 2731 (orange shadow), a contig created during the L. major genome sequence project (Sanger Institute database) but not assembled into the final genome annotation. Also, it is shown in light green the blocks of sequence identity existing between the ends of contigs Cg12141 and Cg12160 and the entry LmjF.24.0770. The chromosomal structure of this region in three Leishmania species (LmjF: L. major; LinJ: L. infantum; LbrM: L. braziliensis) is also shown. b Distribution of RNA-Seq coverage reads and transcripts definition after alignment with the currently annotated genomic region containing the genes LmjF.24.0760 and LmjF.24.0770. c Distribution of RNA-Seq coverage reads and transcripts mapping on the corrected genomic region, indicated in light blue, the new gene (LmjF.24.0765) and transcript (LmjF.24.T0765) uncovered before in this chromosomal region. In red are represented the SIDER2 elements. See legend of Fig. 1 for further explanations. The long shotgun read corresponds with the unassembled contig 2731 (Sanger Institute database)
