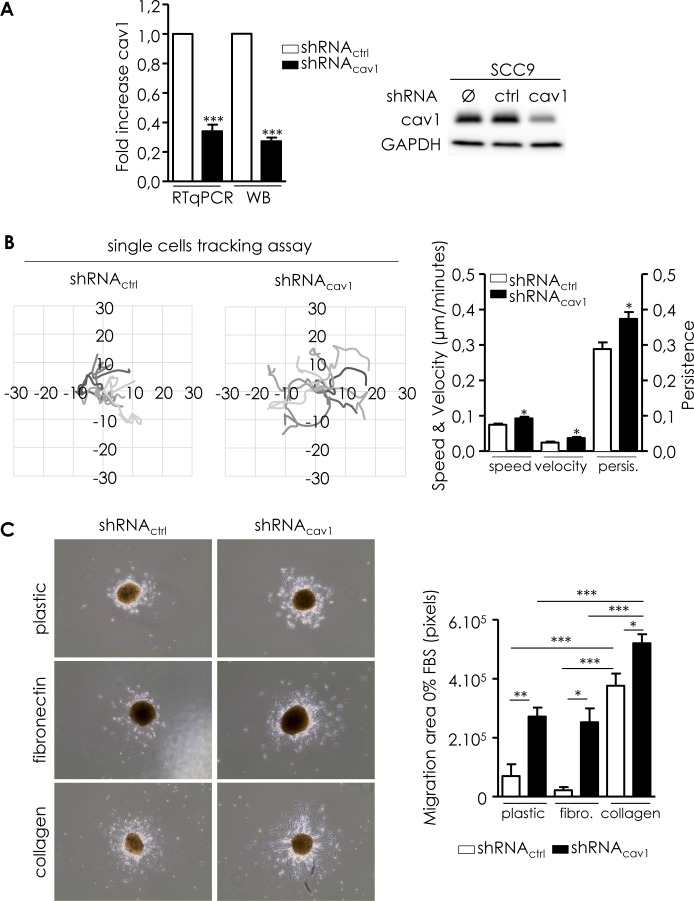
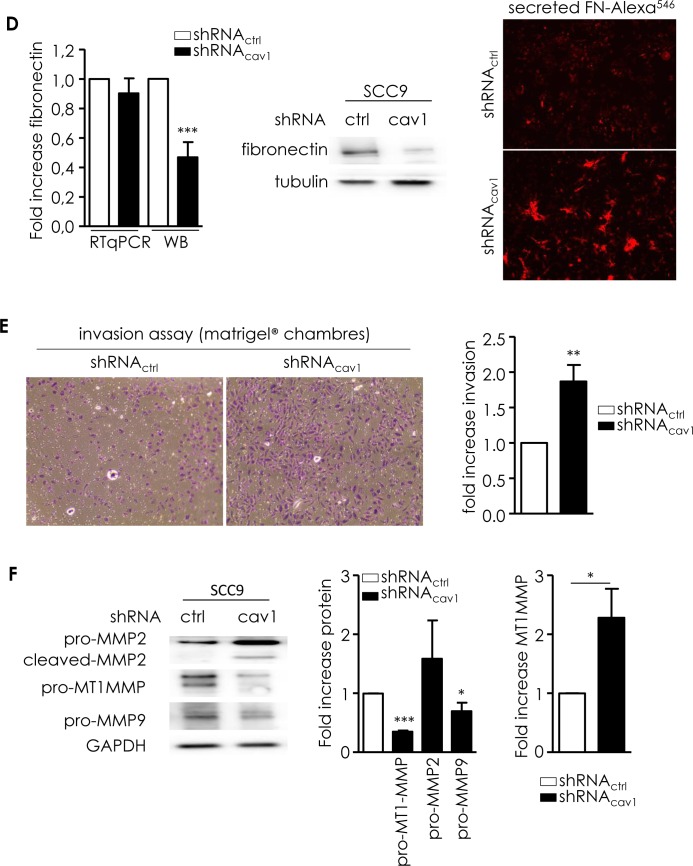
Figure 2. Reduction of Cav1 enables cells motile and invasive properties.
A. Quantitative determination of transcripts and protein expression of Cav1 in shRNActrl- or shRNAcav1-SCC9 cells using qRT-PCR with RNA18S as control and western blot using GAPDH as a loading control. Each bar represents the mean±SEM with ***p < 0.001. B. Analysis of single cell migration of shRNActrl- and shRNAcav1-transfected SCC9. Diagrams represent the migrating trajectories of cells covered during 6 hours by ten representative shRNActrl and shRNAcav1-transfected SCC9. Each bar represents the mean±SEM of the speed, the velocity and the persistence recorded during 6 hours for each cell type (n = 4, *p < 0.05). C. Analysis of collective cell migration of shRNActrl- and shRNAcav1-transfected SCC9. Phase contrast images showing the evasion of shRNActrl- and shRNAcav1-transfected SCC9 from spheroids after 12 hours growth on plastic-, fibronectin- or collagen-coated cells. Each bar represents the mean±SEM area covered by cells evading from the spheroid (n = 7-9, *p < 0.05, **p < 0.01 and ***p < 0.001). D. Left panel: quantitative determination of transcripts and protein expression of fibronectin in shRNActrl- or shRNAcav1-SCC9 cells using qRT-PCR with RNA18S as control and western blot using tubulin as a loading control. Each bar represents the mean±SEM with ***p < 0.001, n = 8. Right panel: immunofluorescence analysis of extracellular secreted fibronectin by shRNActrl- or shRNAcav1-SCC9 cells. Images were taken at 4x magnification with an AXIO Zeiss microscope. E. Phase contrast images showing the invasion of shRNActrl- and shRNAcav1-transfected SCC9 through BioCoat Matrigel® invasion chambers. Cells were stained with crystal violet after 22 hours invasion. Each bar represents the mean±SEM fold increase in invasion (n = 6, **p < 0.01). F. MMPs protein levels were analyzed by western blot analysis using GAPDH as a loading control. Each bar represents the mean±SEM with *p < 0.05 and ***p < 0.001. MT1-MMP was analyzed in shRNActrl- or shRNAcav1-SCC9 cells using qRT-PCR with RNA18S as control. Each bar represents the mean±SEM with *p < 0.05, n = 4.