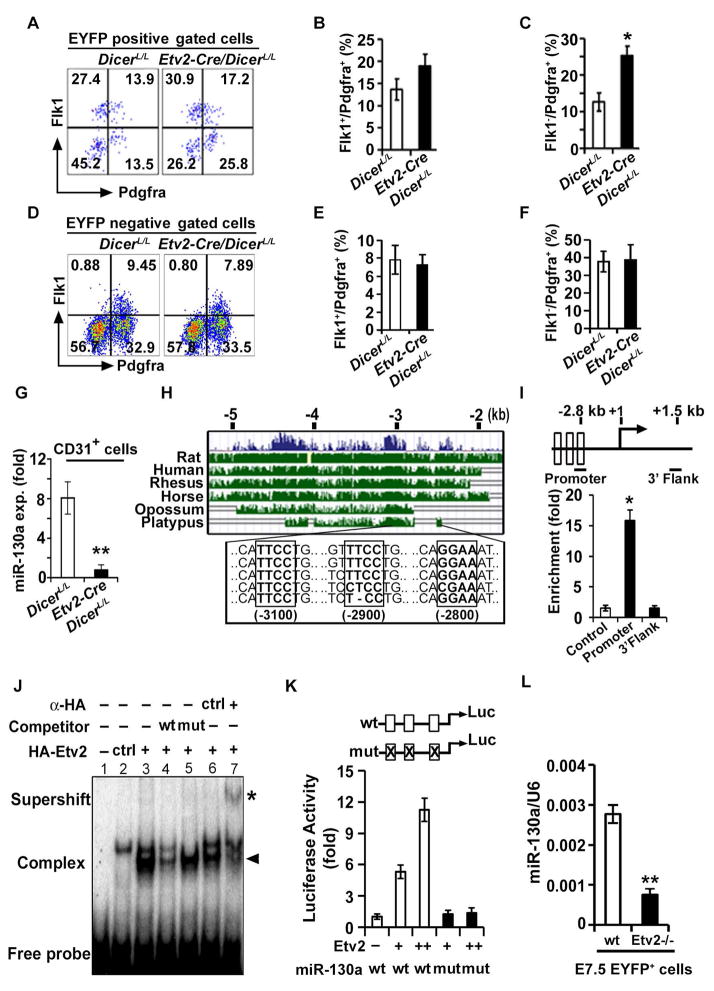
Figure 1. Etv2 modulates miR-130a expression in the endothelial progenitors.
(A–F) Representative FACS profiles (A, D) and quantification (B, C, E, F) of mesodermal populations in Etv2-EYFP:Etv2-Cre;DicerL/L embryos at E7.5 from EYFP+ and EYFP− compartments. (G) qPCR analysis of miR-130a in CD31+ cells sorted from DicerL/L and Etv2-Cre;DicerL/L embryos at E9.5. (H) Evolutionary conservation of the 5.0 kb upstream fragment of the miR-130a gene. (I) Top: Schematic of the 2.8 kb upstream region of the miR-130a promoter. Bottom: ChIP analysis of d4 Dox-inducible HA-Etv2 EBs using an HA antibody. ChIP assay for the Gapdh promoter (Control) and a non-specific locus (miR-130a 3′ UTR region: 3′ Flank) are shown as controls. (J) EMSA showing Etv2 bound to the Ets binding site in the miR-130a promoter region. (K) Luciferase reporter constructs using the miR-130a promoter (-1.0 kb) harboring wild-type (wt; open box) or mutant (mut; crossed box) Etv2 binding sites. (L) qPCR analysis of miR-130a using EYFP+ sorted cells from Etv2 wild-type and mutant embryos at E7.5. Error bars indicate SEM (n = 4; *p<0.05; **p < 0.005) (see also Figure S1).