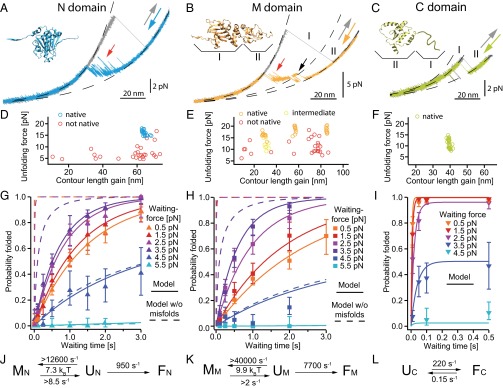
Fig. 2.
Detailed analysis of individual domain constructs. (A−C) Force-extension traces of individual domain constructs (Insets from PDB 2cg9) at a slow pulling speed of 10 nm/s. Unfolding traces (gray) are fitted with WLCs (black dashed). Refolding traces of the N (A, blue) and the M (B, orange) domains populate transient intermediate states (A and B, red arrows). The M domain (B) populates an on-pathway intermediate (black arrow) that corresponds to region II. The C domain (C) shows equilibrium two-state behavior; region I accounts for unstructured regions. More traces are shown in Fig. S2 A, C, and E. (D−F) Scatter plots display unfolding events (unfolding force vs. contour length gain) for the N (D), the M (E), and the C (F) domains. Each scatter plot is derived from 35 to 75 consecutive traces of one molecule. The native structured M domain can appear as a single event (orange, longest length gain) or as a double event (orange, shortest plus medium length gain) depending on the classification algorithm (see SI Methods). If only the on-pathway intermediate is observed (B, region II), it is colored in yellow. (G−I) Averaged refolding probabilities derived from double jump experiments (see Fig. S6 for nonaveraged data) depending on time (y axis) and force (color-coded) for the N (G), the M (H), and the C (I) domains. Probabilities are determined from 11,570 traces (45 molecules), 7,927 traces (14 molecules), and 6,157 traces (9 molecules) for the N, M, and C domains, respectively. The N and M domains show increasing refolding probability with increasing force (G and H, orange to purple markers). Fits are described in J−L. For uncertainties, see SI Methods. (J−L) The atypical refolding behavior of the N and M domains is fully described by a model (J and K) assuming an unfolded state ensemble U that is in equilibrium with fast-forming off-pathway intermediates M, that prevent folding to the native state F. Fitting this model to the nonaveraged data (Fig. S6 A and B) yields fits shown in G and H as continuous lines. Assuming identical folding parameters and neglecting misfolding yields the refolding probabilities shown in G and H as dashed lines. The C domain is fitted with a two-state model assuming an unfolded U and a folded state F in equilibrium (L, continuous lines in I and Fig. S6C). The folding rates at zero force, the equilibrium energy between U and M, and lower estimates for the rates from and to the misfolded states (see Estimate 3) are displayed.